Beyond Single-Model Limits: How Data Fusion Boosts Predictive Accuracy in Biomedicine
This article explores the paradigm shift from single-technique predictive modeling to advanced data fusion strategies, with a focus on applications in biomedical research and drug development.
Beyond Single-Model Limits: How Data Fusion Boosts Predictive Accuracy in Biomedicine
Abstract
This article explores the paradigm shift from single-technique predictive modeling to advanced data fusion strategies, with a focus on applications in biomedical research and drug development. It provides a foundational understanding of core fusion methods—early, late, and intermediate fusion—and delves into their practical implementation for integrating diverse data types like genomic, transcriptomic, proteomic, and clinical data. The content addresses critical challenges such as data heterogeneity and overfitting, offering optimization strategies and theoretical frameworks for method selection. Through rigorous comparative analysis and validation techniques, the article demonstrates how data fusion consistently enhances predictive accuracy, robustness, and generalizability, ultimately paving the way for more reliable tools in precision oncology and therapeutic development.
The Data Fusion Imperative: Moving Beyond Single-Source Predictions
Data fusion, also known as information fusion, is the process of integrating multiple data sources to produce more consistent, accurate, and useful information than that provided by any individual data source [1]. In artificial intelligence (AI), this involves consolidating diverse data types and formats to develop more comprehensive and robust models, leading to enhanced insights and more reliable outcomes [1].
This guide objectively compares the predictive performance of data fusion strategies against single-technique approaches, providing experimental data to underscore the critical trade-offs. The analysis is framed for researchers and professionals who require evidence-based methodologies for improving predictive accuracy in complex domains like drug development.
A Brief History of Data Fusion
The concept of data fusion originated in military operations, where it was initially employed to process information from multiple sources for strategic decision-making and intelligence analysis [1]. This field has since transcended its military origins, evolving in tandem with technological advancements in data acquisition and processing to become a cornerstone of modern AI and data science [1].
Core Data Fusion Methods: A Technical Comparison
Multisource and multimodal data fusion plays a pivotal role in large-scale AI applications. The choice of fusion strategy significantly impacts computational cost and model performance [2]. The three mainstream fusion methods are:
- Early Fusion (Data-Level Fusion): Involves the simple concatenation of original features from different modalities as the input to a single predictive model [2].
- Intermediate Fusion (Feature-Level Fusion): Learns representative features from the original data of each modality first, then fuses these features for predictive classification [2].
- Late Fusion (Decision-Level Fusion): Trains separate models on different data modalities and subsequently fuses the decisions or predictions from these individual models [2].
The workflow and logical relationships between these methods are summarized in the diagram below.
Comparative Performance and Selection Paradigm
Theoretical analysis reveals that no single fusion method is universally superior; the optimal choice depends on data characteristics. Research shows that under a generalized linear model framework, early fusion and late fusion can be mathematically equivalent under specific conditions [2]. However, early fusion can fail when nonlinear relationships exist between features and labels [2].
A critical finding is the existence of a sample size threshold where performance dominance reverses. An approximate equation evaluating the accuracy of early and late fusion as a function of sample size, feature quantity, and modality number enables the creation of a selection paradigm to choose the most appropriate method before task execution, saving computational resources [2].
Experimental Evidence: Data Fusion vs. Single-Source Models
Case Study 1: Genomic and Phenotypic Selection in Plant Breeding
Experimental Protocol: A novel data fusion framework, GPS (Genomic and Phenotypic Selection), was tested for predicting complex traits in crops [3]. The study integrated genomic and phenotypic data through three fusion strategies (data fusion, feature fusion, and result fusion) and applied them to a suite of models, including statistical approaches (GBLUP, BayesB), machine learning models (Lasso, RF, SVM, XGBoost, LightGBM), and deep learning (DNNGP) [3]. The models were rigorously validated on large datasets from four crop species: maize, soybean, rice, and wheat [3].
Performance Comparison:
| Fusion Strategy | Top Model | Accuracy Improvement vs. Best Genomic Model | Accuracy Improvement vs. Best Phenotypic Model |
|---|---|---|---|
| Data Fusion | Lasso_D | +53.4% | +18.7% |
| Feature Fusion | - | Lower than Data Fusion | Lower than Data Fusion |
| Result Fusion | - | Lower than Data Fusion | Lower than Data Fusion |
The Lasso_D model demonstrated exceptional robustness, maintaining high predictive accuracy with sample sizes as small as 200 and showing resilience to variations in genetic marker density [3].
Case Study 2: Sea Surface Nitrate Estimation
Experimental Protocol: This study aimed to enhance the accuracy and spatial resolution of global sea surface nitrate (SSN) retrievals [4]. Researchers developed improved regression and machine learning models that fused satellite-derived data (e.g., sea surface temperature) instead of relying solely on traditional in-situ measurements [4]. The machine learning approach employed seven algorithms: Extremely Randomized Trees (ET), Multilayer Perceptron (MLP), Stacking Random Forest (SRF), Gaussian Process Regression (GPR), Support Vector Machine (SVM), Gradient Boosting Decision Tree (GBDT), and Extreme Gradient Boosting (XGBoost) [4].
Performance Comparison (Root Mean Square Deviation - RMSD):
| Modeling Approach | Key Feature | Best Model | Performance (RMSD μmol/kg) |
|---|---|---|---|
| Regional Empirical Models | Ocean segmented into 5 biome-based regions | - | 1.641 - 2.701 |
| Machine Learning with Data Fusion | Single model for global ocean, no segmentation | XGBoost | 1.189 |
The XGBoost model, which bypassed the need for complex regional segmentation, outperformed all traditional regional empirical models, demonstrating the power of data fusion to create more accurate and universally applicable models [4].
Case Study 3: Hydrocarbon/Imide Ratio Prediction in Oils
Experimental Protocol: Research on oil industry samples compared five data fusion techniques applied to Mid-Infrared (MIR) and Raman spectroscopic data to predict a specific quality parameter (hydrocarbon/imide ratio of an additive) [5]. The fusion techniques were implemented at different levels: low (variable fusion), medium (model result fusion), high, and complex (ensemble learning with spectral data) [5].
Performance Comparison:
| Modeling Technique | Data Source | Relative Result |
|---|---|---|
| Mode A | Single Source (MIR only) | Baseline |
| Mode B | Single Source (Raman only) | Worse than Baseline |
| Mode C | Low-Level Fusion | Better than Baseline |
| Mode D | Intermediate-Level Fusion | Worse than Baseline |
| Mode E | High & Complex-Level Fusion | Best Results |
The results concluded that models using low, high, and complex data fusion techniques yielded better predictions than those built on single-source MIR or Raman data alone [5].
The Scientist's Toolkit: Essential Reagents for Data Fusion
For researchers aiming to implement data fusion frameworks, the following "research reagents" are essential conceptual components.
| Tool/Component | Function in Data Fusion Research |
|---|---|
| Generalized Linear Models (GLMs) | Provides a foundational statistical framework for understanding and deriving equivalence conditions between different fusion methods, particularly early and late fusion [2]. |
| Tree-Based Algorithms (XGBoost, LightGBM, RF) | Highly effective for integrating heterogeneous data sources and modeling complex, nonlinear relationships, often serving as strong baselines or final models in fusion pipelines [3] [4]. |
| Transformer Architectures | Advanced neural networks utilizing self-attention mechanisms to capture long-range dependencies and complex interactions between different data modalities without sequential processing constraints [6]. |
| Shapley Additive exPlanations (SHAP) | A method for interpreting complex, fused models by quantifying the contribution of each input feature (from any source) to the final prediction, ensuring transparency [7] [6]. |
| Partial Least Squares (PLS) Regression | A chemometrics staple used for modeling relationships between two data matrices (e.g., spectral data and quality parameters), frequently employed in low- and mid-level fusion for spectroscopic analysis [5]. |
The body of evidence confirms that data fusion strategies consistently outperform single-technique approaches, delivering substantial gains in predictive accuracy, robustness, and model transferability. The key to success lies in the strategic selection of the fusion method—early, intermediate, or late—which must be guided by the specific data characteristics, sample size, and the linear or nonlinear nature of the problem [2]. For researchers in drug development and beyond, mastering this selection paradigm is no longer a luxury but a necessity for unlocking the next frontier of predictive innovation.
In the pursuit of advanced predictive modeling, researchers and drug development professionals increasingly face a critical choice: whether to rely on single-data sources or integrate multiple modalities through data fusion. This guide examines the core fusion architectures—early, intermediate, and late fusion—that frame this decision within the broader thesis of predictive accuracy versus single-technique research. Technological advancements have generated vast quantities of multi-source heterogeneous data across biomedical domains, from genomic sequences and clinical variables to medical imaging and physiological time series [6] [8]. While single-modality analysis offers simplicity, it fundamentally limits a model's capacity to capture the complex, complementary information distributed across different data types [9].
Multimodal data fusion has emerged as a transformative paradigm to overcome these limitations. By strategically integrating diverse data sources—including clinical records, imaging, molecular profiles, and sensor readings—fusion architectures enable the development of more robust, accurate, and generalizable predictive models [10] [11]. This capability is particularly valuable in precision oncology, where integrating imaging, clinical, and genomic data has demonstrated significant improvements in cancer classification and survival prediction compared to unimodal approaches [9] [12] [8]. The core challenge lies in selecting the optimal fusion strategy that balances model complexity with predictive performance while accounting for domain-specific constraints.
This guide provides a systematic comparison of the three principal fusion architectures, supported by experimental data and methodological protocols. By objectively evaluating the performance of each fusion type against single-technique alternatives, we aim to equip researchers with the evidence needed to make informed decisions in their predictive modeling workflows.
Fusion Architecture Fundamentals
Definitions and Conceptual Frameworks
The three primary fusion strategies—early, intermediate, and late fusion—differ fundamentally in their approach to integrating information across modalities, each with distinct technical implementations and performance characteristics.
Early Fusion (Data-Level Fusion): This approach involves concatenating raw or preprocessed data from different modalities into a unified input vector before feeding it into a single model [2] [13]. For example, in biomedical applications, clinical variables, genomic data, and imaging features might be combined into one comprehensive input matrix. The defining characteristic of early fusion is its ability to learn complex interactions between modalities from the outset, as expressed by the generalized linear model formulation:
g_E(μ) = η_E = Σ(w_i * x_i)where all featuresx_ifrom different modalities are combined with weight coefficientsw_i[2].Intermediate Fusion (Feature-Level Fusion): This strategy processes each modality through separate feature extractors before combining the learned representations at an intermediate layer within the model architecture [10] [11]. Also known as joint fusion or model-level fusion, this approach preserves modality-specific characteristics while learning cross-modal interactions through specialized fusion layers. Intermediate fusion effectively balances the preservation of modality-specific features with the learning of joint representations, making it particularly valuable for capturing complex inter-modal relationships in biomedical contexts [10].
Late Fusion (Decision-Level Fusion): This method trains separate models on each modality independently and combines their predictions at the decision stage through a fusion function [12] [2] [8]. The mathematical formulation can be expressed as training sub-models for each modality:
g_Lk(μ) = η_Lk = Σ(w_jk * x_jk)fork = 1,2,...,Kmodalities, then aggregating the outputs through a fusion function:output_L = f(g_L1^{-1}(η_L1), g_L2^{-1}(η_L2), ..., g_LK^{-1}(η_LK))[2]. This approach offers strong resistance to overfitting, especially with imbalanced modality dimensionalities [8].
Architectural Workflows
The diagram below illustrates the fundamental workflow differences between the three core fusion architectures.
Performance Comparison: Fusion vs. Single-Modality Approaches
Quantitative Performance Analysis
Experimental evidence across multiple biomedical domains demonstrates that multimodal fusion approaches consistently outperform single-modality methods, though the optimal fusion strategy varies by application context and data characteristics.
Table 1: Comparative Performance of Fusion Architectures Across Biomedical Domains
| Application Domain | Single-Modality Baseline (Performance) | Early Fusion (Performance) | Intermediate Fusion (Performance) | Late Fusion (Performance) | Optimal Fusion Strategy |
|---|---|---|---|---|---|
| Breast Cancer Survival Prediction [12] | Clinical data only (C-index: 0.76) | N/R | N/R | Clinical + Omics (C-index: 0.89) | Late Fusion |
| Chemical Engineering Project Prediction [6] | Traditional ML (Accuracy: ~71.6%) | N/R | Standard Transformer (Accuracy: ~84.9%) | N/R | Improved Transformer (Accuracy: 91.0%) |
| Prostate Cancer Classification [9] | Unimodal approaches (AUC: <0.85) | Common with CNNs (AUC: ~0.82-0.88) | Varied performance | Less common | Early/Intermediate Fusion |
| Multi-Omics Cancer Survival [8] | Multiple unimodal baselines | Lower vs. late fusion | Moderate performance | Consistently superior | Late Fusion |
| Biomedical Time Series [11] | Unimodal deep learning | Limited by misalignment | Superior accuracy & robustness | Moderate performance | Intermediate Fusion |
Table 2: Contextual Advantages and Limitations of Fusion Architectures
| Fusion Type | Key Advantages | Primary Limitations | Optimal Application Context |
|---|---|---|---|
| Early Fusion | Captures complex feature interactions immediately; Single model simplicity | Vulnerable to overfitting with high-dimensional data; Requires modality alignment | Low-dimensional, aligned modalities; Strong inter-modal correlations |
| Intermediate Fusion | Balances specificity and interaction; Flexible architecture design | Complex model design; Higher computational demand | Cross-modal relationship learning; Handling temporal misalignment |
| Late Fusion | Robust to overfitting; Handles data heterogeneity; Enables modality weighting | Cannot learn cross-modal interactions; Requires sufficient unimodal data | High-dimensional, heterogeneous data; Independent modality distributions |
Theoretical Performance Under varying Conditions
Research by [2] provides mathematical analysis of the conditions under which different fusion strategies excel, proposing an approximate equation for evaluating the accuracy of early and late fusion as a function of sample size, feature quantity, and modality number. Their work identifies a critical sample size threshold at which the performance dominance of early and late fusion models reverses:
- With small sample sizes relative to feature dimensions, late fusion consistently outperforms early fusion due to its resistance to overfitting [2].
- Early fusion theoretically outperforms late fusion in binary classification problems given a perfect model and sufficiently large sample size [2].
- The presence of nonlinear feature-label relationships can cause early fusion to fail, making late or intermediate fusion more appropriate for complex, nonlinear biomedical data [2].
Experimental Protocols and Methodologies
Benchmarking Fusion Architectures in Survival Prediction
The experimental protocol for comparing fusion architectures in cancer survival prediction [12] [8] follows a rigorous methodology:
Data Collection and Preprocessing: Aggregate multi-omics data (transcripts, proteins, metabolites), clinical variables, and histopathology images from curated sources like The Cancer Genome Atlas (TCGA). Implement appropriate normalization, batch effect correction, and missing data imputation.
Feature Selection and Dimensionality Reduction: Apply modality-specific feature selection to address high dimensionality. Common approaches include:
Model Training with Multiple Fusion Strategies:
- Early Fusion: Concatenate selected features from all modalities into a unified input matrix for a single survival model.
- Intermediate Fusion: Process each modality through separate subnetworks, then combine features using attention mechanisms or concatenation before the final prediction layer [10].
- Late Fusion: Train separate survival models for each modality, then combine predictions through weighted averaging or meta-learners [12] [8].
Evaluation and Validation: Assess performance using concordance index (C-index) with confidence intervals from multiple training-test splits, accounting for considerable uncertainty from different data partitions [8]. Additional evaluation includes calibration analysis and time-dependent AUC metrics.
Domain-Specific Implementation Variations
Across different biomedical applications, the core experimental protocol adapts to domain-specific requirements:
Biomedical Time Series Prediction [11]: Incorporates temporal alignment mechanisms and specialized architectures (LSTMs, Transformers) to handle varying sampling rates across physiological signals, clinical events, and medication records.
Medical Imaging Integration [14]: Employs convolutional neural networks (CNNs) for feature extraction from imaging data, followed by radiomics feature selection and habitat imaging analysis before fusion with clinical and genomic data.
Chemical Engineering Applications [6]: Utilizes improved Transformer architectures with multi-scale attention mechanisms to handle vastly different temporal hierarchies, from millisecond sensor readings to monthly progress reports.
The diagram below illustrates a comprehensive experimental workflow for comparing fusion methodologies in biomedical research.
Successful implementation of fusion architectures requires careful selection of computational frameworks, data resources, and evaluation tools. The following table details essential components for constructing and validating multimodal fusion models.
Table 3: Essential Research Reagents and Computational Resources for Fusion Experiments
| Resource Category | Specific Tools & Datasets | Primary Function | Application Context |
|---|---|---|---|
| Public Data Repositories | The Cancer Genome Atlas (TCGA) [12] [8] | Provides curated multi-omics, clinical, and imaging data for cancer research | Benchmarking fusion models across diverse cancer types |
| PeMS Traffic Data [15] | Offers structured temporal data for long-term prediction validation | Testing fusion approaches on heterogeneous time series | |
| Computational Frameworks | Transformer Architectures [6] | Handles variable-length sequences and captures long-range dependencies | Processing data with vastly different sampling frequencies |
| Adaptive Multimodal Fusion Networks (AMFN) [11] | Dynamically captures inter-modal dependencies with attention mechanisms | Biomedical time series with misaligned modalities | |
| Evaluation Metrics | Concordance Index (C-index) [12] [8] | Evaluates ranking accuracy of survival predictions | Assessing clinical prediction models |
| SHAP/LIME Analysis [14] | Provides model interpretability and feature importance | Understanding fusion model decisions for clinical translation | |
| Fusion-Specific Libraries | AZ-AI Multimodal Pipeline [8] | Python library for multimodal feature integration and survival prediction | Streamlining preprocessing, fusion, and evaluation workflows |
The evidence consistently demonstrates that multimodal fusion architectures significantly outperform single-modality approaches across diverse biomedical applications, with performance improvements of 6-20% depending on the domain and data characteristics [6] [12] [8]. However, the optimal fusion strategy is highly context-dependent, requiring careful consideration of data properties and application requirements.
For researchers and drug development professionals, the following strategic guidelines emerge from experimental evidence:
Prioritize Late Fusion when working with high-dimensional data, limited samples, or significant heterogeneity across modalities, particularly in survival prediction and multi-omics integration [12] [8].
Select Intermediate Fusion when capturing complex cross-modal relationships is essential, and sufficient data exists to train more sophisticated architectures, especially for biomedical time series and imaging applications [10] [11].
Consider Early Fusion primarily for low-dimensional, well-aligned modalities where capturing fine-grained feature interactions is critical to prediction performance [2] [13].
Implement Rigorous Evaluation practices including multiple data splits, confidence intervals for performance metrics, and comparisons against unimodal baselines to ensure meaningful conclusions about fusion effectiveness [8].
The strategic integration of multimodal data through appropriate fusion architectures represents a substantial advancement over single-technique research, offering enhanced predictive accuracy and more robust models for critical applications in drug development and precision medicine. As fusion methodologies continue to evolve, particularly with advances in attention mechanisms and transformer architectures, their capacity to translate heterogeneous data into actionable insights will further expand, solidifying their role as essential tools in biomedical research.
In predictive research, single-source data or unimodal models often struggle with inherent uncertainties, including high dimensionality, low signal-to-noise ratios, and data heterogeneity. These challenges are particularly acute in high-stakes fields like drug development and precision oncology, where accurate predictions directly impact patient outcomes and therapeutic discovery. Data fusion has emerged as a transformative methodology that systematically mitigates these uncertainties by integrating complementary information from multiple sources, features, or models. The core theoretical advantage of fusion lies in its ability to synthesize disparate evidence, thereby reducing variance, counteracting biases in individual data sources, and producing more robust and reliable predictive outputs. This review synthesizes current evidence and theoretical frameworks demonstrating how fusion techniques enhance predictive accuracy and reliability compared to single-modality approaches, with particular emphasis on applications relevant to biomedical research and pharmaceutical development.
Theoretical Foundations: How Fusion Mitigates Uncertainty
Core Mechanisms of Uncertainty Reduction
Data fusion reduces uncertainty through several interconnected theoretical mechanisms. First, it addresses the complementarity principle, where different data modalities capture distinct aspects of the underlying system. For instance, in cancer survival prediction, genomic data may reveal mutational drivers, clinical variables provide physiological context, and proteomic measurements offer functional insights. Fusion integrates these complementary perspectives, creating a more complete representation of the phenomenon than any single source can provide [8]. Second, fusion leverages information redundancy when multiple sources provide overlapping evidence about the same underlying factor. This redundancy allows the system to cross-validate information, reducing the impact of noise and measurement errors in individual sources while increasing overall confidence in the consolidated output [2] [8].
The mathematical relationship between data fusion and uncertainty reduction can be conceptualized through statistical learning theory. In late fusion, for example, where predictions from multiple unimodal models are aggregated, the variance of the fused prediction can be substantially lower than that of individual models, particularly when the errors between models are uncorrelated. This variance reduction directly decreases prediction uncertainty and enhances generalizability, especially valuable when working with limited sample sizes common in biomedical research [2] [8].
Categorical Framework of Fusion Techniques
Fusion methodologies are systematically categorized based on the stage at which integration occurs, each with distinct theoretical properties affecting uncertainty reduction:
- Early Fusion (Data-Level Fusion): Raw data from multiple sources are concatenated before feature extraction or model training. This approach preserves potential interactions between raw data sources but risks increased dimensionality and overfitting, particularly with small sample sizes [2] [8].
- Intermediate Fusion (Feature-Level Fusion): Features are first extracted from each data source independently, then integrated into a shared representation space. This balances specificity with joint learning, allowing the model to discover complex cross-modal interactions while maintaining some modality-specific processing [2].
- Late Fusion (Decision-Level Fusion): Separate models are trained on each data source, and their predictions are aggregated. This approach is particularly robust to data heterogeneity and missing modalities, as it accommodates different model architectures tailored to each data type [2] [8].
Table 1: Comparison of Fusion Strategies and Their Properties
| Fusion Type | Integration Stage | Theoretical Advantages | Common Applications |
|---|---|---|---|
| Early Fusion | Raw data input | Preserves cross-modal interactions; Maximizes theoretical information | Multimodal data with strong interconnections; Large sample sizes |
| Intermediate Fusion | Feature representation | Balances specificity and joint learning; Handles some heterogeneity | Hierarchical data structures; Moderate sample sizes |
| Late Fusion | Model predictions | Robust to data heterogeneity; Resistant to overfitting | High-dimensional data with small samples; Missing modalities |
Experimental Evidence: Quantitative Comparisons of Fusion Performance
Fusion in Healthcare and Disease Prediction
Substantial empirical evidence demonstrates fusion's superiority over single-modality approaches across healthcare domains. A fusion-based machine learning approach for diabetes identification, combining Support Vector Machine (SVM) and Artificial Neural Network (ANN) classifiers, achieved a 94.67% prediction accuracy, exceeding the performance of either classifier alone and outperforming the best previously reported model by approximately 1.8% [16]. This fusion architecture specifically enhanced both sensitivity (89.23%) and specificity (97.32%), indicating more reliable classification across different patient subgroups [16].
In oncology, a comprehensive machine learning pipeline for multimodal data fusion analyzed survival prediction in cancer patients using The Cancer Genome Atlas (TCGA) data, incorporating transcripts, proteins, metabolites, and clinical factors [8]. The research demonstrated that late fusion models "consistently outperformed single-modality approaches in TCGA lung, breast, and pan-cancer datasets, offering higher accuracy and robustness" [8]. This performance advantage was particularly pronounced given the challenging characteristics of biomedical data, including high dimensionality, small sample sizes, and significant heterogeneity across modalities.
Cross-Domain Validation of Fusion Efficacy
Beyond healthcare, fusion methods demonstrate similar advantages in diverse predictive domains. In financial market prediction, fusion techniques that integrate numerical data with textual information from news and social media have shown "substantial improvements in profit" and forecasting accuracy [17]. A systematic review of fusion techniques in this domain between 2016-2025 highlights how integrating disparate data sources enhances prediction reliability by capturing both quantitative market data and qualitative sentiment indicators [17].
In chemical engineering construction projects, an improved Transformer architecture for multi-source heterogeneous data fusion achieved "prediction accuracies exceeding 91% across multiple tasks," representing improvements of "up to 19.4% over conventional machine learning techniques and 6.1% over standard Transformer architectures" [6]. This approach successfully integrated structured numerical measurements, semi-structured operational logs, and unstructured textual documentation, demonstrating fusion's capacity to handle extreme data heterogeneity while reducing predictive uncertainty.
Table 2: Quantitative Performance Comparison Across Domains
| Application Domain | Single-Model Performance | Fusion Approach Performance | Performance Gain | Key Fusion Method |
|---|---|---|---|---|
| Diabetes Identification | 92.8% (DELM) [16] | 94.67% [16] | +1.87% | Classifier Fusion (SVM+ANN) |
| Cancer Survival Prediction | Variable by modality [8] | Consistently superior [8] | Significant | Late Fusion |
| Chemical Engineering Project Management | ~76% (Conventional ML) [6] | 91%+ [6] | +15% | Transformer-based Fusion |
| Financial Market Forecasting | Baseline single-source [17] | Substantially improved profit [17] | Significant | Multimodal Text-Data Fusion |
Methodological Protocols: Implementing Fusion Strategies
Experimental Workflow for Fusion Analysis
The implementation of effective fusion strategies follows systematic methodological protocols. The AstraZeneca–artificial intelligence (AZ-AI) multimodal pipeline for survival prediction in cancer patients provides a representative framework for fusion implementation [8]. This pipeline encompasses data preprocessing, multiple fusion strategies, diverse feature reduction approaches, and rigorous evaluation metrics, offering a standardized methodology for comparing fusion efficacy against unimodal benchmarks.
Diagram 1: Experimental Workflow for Multimodal Data Fusion
Fusion Selection Paradigm and Critical Thresholds
Research has established theoretical frameworks to guide the selection of appropriate fusion strategies based on dataset characteristics. A comparative analysis of three data fusion methods proposed a "critical sample size threshold at which the performance dominance of early fusion and late fusion models undergoes a reversal" [2]. This paradigm enables researchers to select the optimal fusion approach before task execution, improving computational efficiency and predictive performance.
The theoretical analysis demonstrates that under generalized linear models, early and late fusion achieve equivalence under specific mathematical conditions, but early fusion may fail when nonlinear feature-label relationships exist across modalities [2]. This work further provides an "approximate equation for evaluating the accuracy of early and late fusion methods as a function of sample size, feature quantity, and modality number" [2], offering a principled basis for fusion strategy selection rather than relying solely on empirical comparisons.
Implementation Toolkit: Research Reagent Solutions for Data Fusion
Successful implementation of fusion strategies requires both computational frameworks and analytical methodologies. The following toolkit outlines essential components for developing and evaluating fusion-based predictive systems.
Table 3: Essential Research Reagents for Fusion Implementation
| Tool/Component | Category | Function in Fusion Research | Representative Examples |
|---|---|---|---|
| Multimodal Pipeline Architecture | Computational Framework | Standardizes preprocessing, fusion strategies, and evaluation for reproducible comparison | AZ-AI Multimodal Pipeline [8] |
| Feature Selection Methods | Analytical Method | Reduces dimensionality while preserving predictive signals; mitigates overfitting | Pearson/Spearman correlation, Mutual Information [8] |
| Hybrid Validation Protocols | Evaluation Framework | Combines cross-validation with sampling methods to assess generalizability | Fusion Sampling Validation (FSV) [18] |
| Transformer Architectures | Modeling Framework | Handles heterogeneous data types through unified embeddings and attention mechanisms | Improved Transformer with multi-scale attention [6] |
| Ensemble Survival Models | Predictive Modeling | Integrates multiple survival models for more robust time-to-event predictions | Gradient boosting, random forests [8] |
Diagram 2: Architectural Comparison of Early vs. Late Fusion Strategies
The theoretical principles and empirical evidence consolidated in this review demonstrate that data fusion provides systematic advantages for reducing predictive uncertainty and increasing reliability across multiple domains, particularly in biomedical and pharmaceutical applications. The performance gains observed in diabetes identification, cancer survival prediction, and drug development contexts consistently show that strategically integrated multimodal information outperforms single-source approaches. The critical theoretical insight is that fusion mitigates the limitations and uncertainties inherent in individual data sources by leveraging complementarity, redundancy, and error independence across modalities.
For researchers and drug development professionals, these findings underscore the importance of adopting fusion methodologies in predictive modeling workflows. The availability of standardized pipelines, theoretical selection frameworks, and specialized architectures like Transformers for heterogeneous data makes fusion increasingly accessible for practical implementation. As multimodal data generation continues to accelerate in life sciences, fusion approaches will become increasingly essential for extracting reliable insights, reducing decision uncertainty, and advancing precision medicine initiatives. Future research directions include developing more sophisticated cross-modal alignment techniques, adaptive fusion mechanisms that dynamically weight source contributions based on quality and relevance, and enhanced interpretability frameworks to build trust in fused predictive systems.
The modern scientific and business landscapes are defined by an explosion of data, generated from a proliferating number of disparate sources. In this context, data integration—the process of combining and harmonizing data from multiple sources, formats, or systems into a unified, coherent dataset—has transitioned from a technical convenience to a strategic necessity [19]. For researchers, scientists, and drug development professionals, this is not merely an IT challenge but a fundamental component of accelerating discovery and enhancing predictive accuracy. The central thesis of this guide is that integrated data solutions, particularly through advanced fusion methods, consistently demonstrate superior predictive performance compared to single-technique or single-source approaches. This is evidenced by a growing body of research across fields from genomics to clinical drug development, where the fusion of disparate data modalities is unlocking new levels of insight, robustness, and transferability in predictive models [3] [20]. This guide objectively compares the performance of integrated data solutions against traditional alternatives, providing the detailed experimental data and methodologies needed to inform the selection of tools and frameworks for high-stakes research environments.
The Integrated Data Landscape: Tools and Platforms
The market for data integration tools is diverse, with platforms engineered for specific use cases such as analytics, operational synchronization, or enterprise-scale ETL (Extract, Transform, Load). The choice of tool is critical and should be driven by the primary intended outcome [21].
Table 1: Comparison of Data Integration Tool Categories
| Category | Representative Tools | Core Strength | Ideal Outcome |
|---|---|---|---|
| Modern ELT for Analytics | Fivetran, Airbyte, Estuary [19] [21] [22] | Reliably moving data from sources to a central data warehouse for analysis [21]. | Dashboards, AI/ML features, historical analysis [21]. |
| Real-Time Operational Sync | Stacksync [21] | Maintaining real-time, bi-directional data consistency across live operational systems (e.g., CRM, ERP) with conflict resolution [21]. | Operational consistency, accurate customer records, synchronized orders [21]. |
| Enterprise ETL & iPaaS | Informatica PowerCenter, MuleSoft, Talend [19] [21] | Handling complex, high-volume data transformation and integration requirements in large IT environments [19] [21]. | Complex data workflows, application networking, large-scale batch processing. |
Selecting the right platform involves evaluating key technical criteria. Connectivity is paramount; the tool must offer pre-built connectors to your necessary data sources and APIs [19]. Scalability ensures the platform can handle growing data volumes, while data quality and governance capabilities like profiling, cleansing, and lineage tracking are essential for research integrity [19] [23]. Finally, the movement model—whether it's batch ETL, ELT (Extract, Load, Transform), or real-time Change Data Capture (CDC)—must align with the latency requirements of your projects [21] [22].
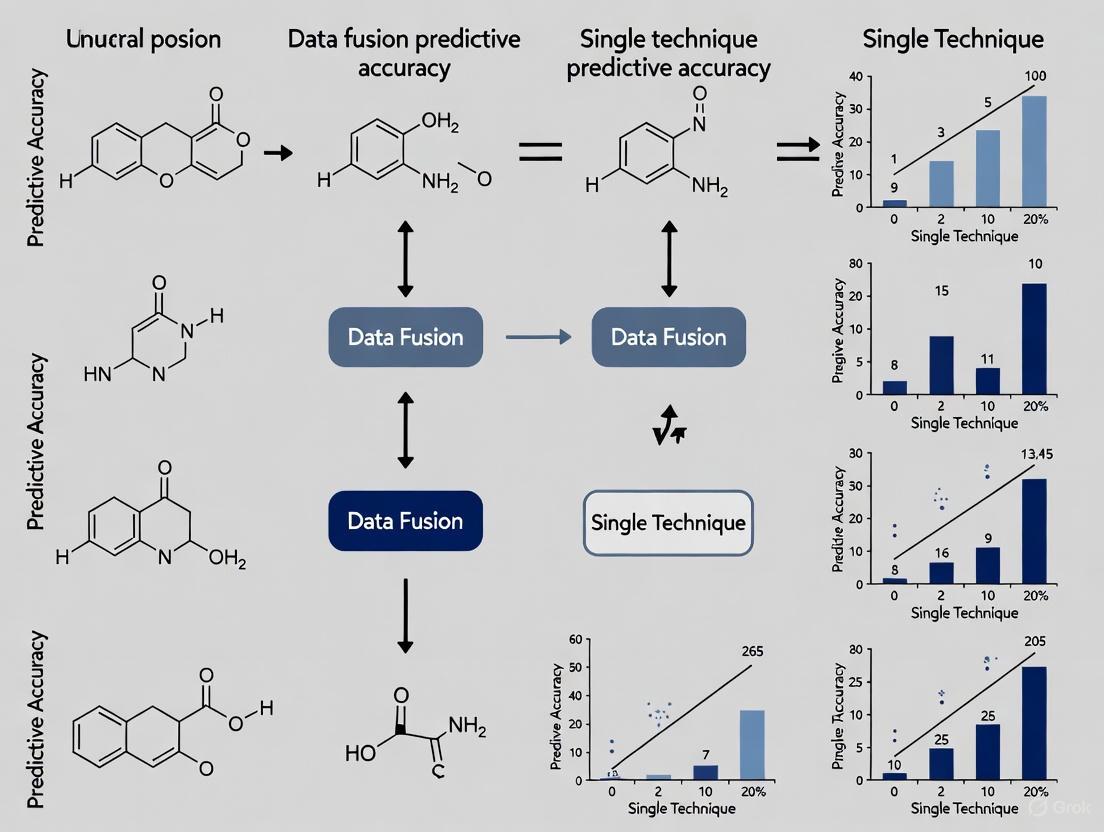
Data Fusion vs. Single-Technique Approaches: Experimental Evidence
The superiority of integrated data solutions is not merely theoretical but is being rigorously demonstrated through controlled experiments across multiple scientific domains. The following section summarizes key experimental findings and protocols that directly compare fused data approaches against single-modality baselines.
Evidence from Genomic and Phenotypic Selection
A groundbreaking study in plant science introduced the GPS framework, a novel data fusion strategy for integrating genomic and phenotypic data to improve genomic selection (GS) and phenotypic selection (PS) for complex traits [3]. The researchers systematically compared three fusion strategies—data fusion, feature fusion, and result fusion—against the best single-technique models across four crop species (maize, soybean, rice, and wheat).
Table 2: Predictive Accuracy in Genomic and Phenotypic Selection [3]
| Model Type | Specific Model | Key Performance Finding | Comparative Improvement |
|---|---|---|---|
| Best Single-Technique (GS) | LightGBM | Baseline accuracy for Genomic Selection | -- |
| Best Single-Technique (PS) | Lasso | Baseline accuracy for Phenotypic Selection | -- |
| Data Fusion Model | Lasso_D | Achieved the highest accuracy of all tested models | 53.4% higher than best GS model (LightGBM); 18.7% higher than best PS model (Lasso) [3]. |
Experimental Protocol for Genomic-Phenotypic Fusion [3]:
- Datasets: Large-scale datasets from four crop species: maize, soybean, rice, and wheat.
- Models: A suite of models was employed, including statistical approaches (GBLUP, BayesB), machine learning models (Lasso, RF, SVM, XGBoost, LightGBM), a deep learning method (DNNGP), and a phenotype-assisted model (MAK).
- Fusion Strategies:
- Data Fusion: Raw genomic and phenotypic data were concatenated into a single input vector for model training.
- Feature Fusion: Features were learned from genomic and phenotypic data separately and then fused for predictive classification.
- Result Fusion: Separate models were trained on genomic and phenotypic data, and their decisions were aggregated for a final prediction.
- Evaluation: Predictive accuracy for complex traits was the primary metric. The robustness of the top-performing model (Lasso_D) was further tested with varying sample sizes and SNP densities, and its transferability was assessed using multi-environmental data.
Evidence from Chemical Engineering and Multi-Modal Prediction
Research in chemical engineering construction further validates the advantages of integrated data solutions. A 2025 study developed a framework for multi-source heterogeneous data fusion using an improved Transformer architecture to integrate diverse data types, including structured numerical measurements, semi-structured operational logs, and unstructured textual documentation [6].
Table 3: Predictive Modeling Accuracy in Chemical Engineering [6]
| Modeling Approach | Reported Prediction Accuracy | Comparative Performance |
|---|---|---|
| Conventional Machine Learning | Baseline | Up to 19.4% lower than the proposed fusion model |
| Standard Transformer Architecture | Baseline | 6.1% lower than the improved fusion model |
| Improved Transformer with Multi-Source Data Fusion | Exceeded 91% across multiple tasks (progress, quality, risk) | Represented an improvement of up to 19.4% over conventional ML and 6.1% over standard Transformers [6]. |
Experimental Protocol for Multi-Modal Transformer Fusion [6]:
- Data: Multi-source heterogeneous data from chemical engineering construction projects, including sensor readings, progress reports, and material logs.
- Model Architecture: An improved Transformer model with a domain-specific multi-scale attention mechanism was used. This mechanism explicitly models temporal hierarchies to handle data streams with vastly different sampling frequencies.
- Key Innovations:
- Contrastive Cross-Modal Alignment: This framework learns semantic correspondences between heterogeneous modalities without manually crafted feature mappings.
- Adaptive Weight Allocation: This algorithm dynamically adjusts the contribution of each data source based on real-time quality assessment and task-specific relevance.
- Evaluation: The model was simultaneously evaluated on progress estimation, quality assessment, and risk evaluation tasks. It also demonstrated robust anomaly detection (exceeding 92% detection rates) and real-time processing performance (under 200 ms).
Theoretical Underpinnings of Fusion Performance
The empirical results are supported by theoretical analyses that explain why and when different fusion methods excel. A comparative analysis of early, late, and gradual fusion methods derived equivalence conditions between early and late fusion within generalized linear models [2]. Crucially, the study proposed an equation to evaluate the accuracy of early and late fusion as a function of sample size, feature quantity, and modality number, identifying a critical sample size threshold at which the performance dominance of early and late fusion models reverses [2]. This provides a principled basis for selecting a fusion method prior to task execution, moving beyond reliance on experimental comparisons alone.
Visualization of Data Fusion Methodologies
The following diagrams illustrate the core architectures and workflows of the data fusion methods discussed in the experimental evidence.
Conceptual Data Fusion Workflow
Technical Fusion Strategies
The Scientist's Toolkit: Essential Reagents & Platforms
For researchers embarking on data integration projects, the following tools and platforms constitute the modern "reagent kit" for building robust, integrated data solutions.
Table 4: Essential Research Reagent Solutions for Data Integration
| Tool / Solution | Category / Function | Brief Explanation & Research Application |
|---|---|---|
| Fivetran [19] [21] | Automated ELT Pipeline | A fully managed service for automating data movement from sources to a data warehouse. Ideal for analytics teams needing reliable, hands-off data ingestion for downstream analysis. |
| Airbyte [21] [22] | Open-Source ELT | Provides flexibility and cost-effectiveness with a large library of connectors. Suited for technical teams requiring control over their data pipelines and wishing to avoid vendor lock-in. |
| Informatica PowerCenter [19] [21] | Enterprise ETL | A robust, scalable solution for complex, high-volume data transformation workflows. Meets the needs of large enterprises in regulated industries like healthcare and finance. |
| Estuary [22] | Real-Time ELT/CDC | Supports both real-time and batch data integration with built-in data replay. Fits projects requiring low-latency data capture and synchronization for real-time analytics. |
| Stacksync [21] | Real-Time Operational Sync | A platform engineered for bi-directional synchronization between live systems (e.g., CRM ERP). Solves the problem of operational data inconsistency with conflict resolution. |
| Transformer Architectures [6] | Multi-Modal ML Framework | Deep learning models, particularly those with enhanced attention mechanisms, are pivotal for fusing heterogeneous data (text, numerical, sequential) into a unified representation for prediction. |
| Causal Machine Learning (CML) [20] | Causal Inference | Methods like targeted maximum likelihood estimation and doubly robust inference integrate RCT data with Real-World Data (RWD) to estimate causal treatment effects and identify patient subgroups. |
The collective evidence from genomics, chemical engineering, and clinical science presents a compelling case: the future of predictive modeling and scientific discovery is inextricably linked to the ability to effectively integrate and fuse disparate data sources. The experimental data consistently shows that integrated data solutions can achieve significant improvements in accuracy—ranging from 6.1% to over 50%—compared to single-technique approaches [3] [6]. This performance gain is coupled with enhanced robustness, transferability, and the ability to model complex, real-world systems more faithfully. For the modern researcher, proficiency with the tools and methodologies of data fusion is no longer a niche skill but a core component of the scientific toolkit, essential for driving innovation and achieving reliable, impactful results.
Fusion in Action: Methodologies for Integrating Multimodal Biomedical Data
In the evolving landscape of data-driven research, the limitations of unimodal analysis have become increasingly apparent. Data fusion, the process of integrating multiple data sources to produce more consistent, accurate, and useful information than that provided by any individual source, has emerged as a critical methodology across scientific disciplines [24]. For researchers and drug development professionals, selecting an appropriate fusion strategy is paramount, as it directly impacts the predictive accuracy and reliability of analytical models. This guide provides a comparative analysis of the three primary fusion levels—data-level (early), feature-level (intermediate), and decision-level (late) fusion—framed within the broader thesis of how multimodal integration enhances predictive performance compared to single-technique approaches.
The fundamental premise of data fusion is that different data sources often provide complementary information [24]. In a drug development context, this could involve integrating genomic data, clinical variables, and high-resolution imaging to achieve a more comprehensive understanding of disease mechanisms or treatment effects. The strategic combination of these disparate data types can reveal interactions and patterns that remain obscured when modalities are analyzed independently.
Theoretical Foundations and Classification
Data fusion techniques are systematically classified based on the processing stage at which integration occurs. The most prevalent framework in clinical and scientific research distinguishes between data-level, feature-level, and decision-level fusion [25] [24]. Data-level fusion (also called early fusion) combines raw data from multiple sources before feature extraction [26]. Feature-level fusion (intermediate fusion) integrates processed features extracted from each modality separately [25]. Decision-level fusion (late fusion) combines outputs from multiple classifiers, each trained on different data modalities [2].
A more detailed conceptual model, established by the Joint Directors of Laboratories (JDL), further categorizes data fusion into five progressive levels: source preprocessing (Level 0), object refinement (Level 1), situation assessment (Level 2), impact assessment (Level 3), and process refinement (Level 4) [24]. This multi-layered approach facilitates increasingly sophisticated inferences, moving from basic data correlation to high-level impact assessment and resource management.
Table 1: Classification Frameworks for Data Fusion
| Classification Basis | Categories | Description | Relevant Context |
|---|---|---|---|
| Abstraction Level [24] | Data/Input-Level | Fusion of raw or minimally processed data | Combines sensor readings or raw images |
| Feature-Level | Fusion of extracted features | Merges shape, texture, or statistical features | |
| Decision-Level | Fusion of model outputs or decisions | Averages or weights classifier predictions | |
| Dasarathy's Model [24] | Data In - Data Out (DAI-DAO) | Input & output are raw data | Signal/image processing algorithms |
| Data In - Feature Out (DAI-FEO) | Input is raw data, output is features | Feature extraction techniques | |
| Feature In - Feature Out (FEI-FEO) | Input & output are features | Intermediate-level fusion | |
| Feature In - Decision Out (FEI-DEO) | Input is features, output is decisions | Standard classification systems | |
| Decision In - Decision Out (DEI-DEO) | Input & output are decisions | Ensemble methods, model stacking | |
| Data Relationships [24] | Complementary | Data represents different parts of a scene | Non-overlapping sensor fields of view |
| Redundant | Data provides overlapping information | Multiple sensors measuring same parameter | |
| Cooperative | Data combined into new, more complex information | Multi-modal (e.g., audio-video) fusion |
The theoretical relationship between early (data-level) and late (decision-level) fusion has been formally explored within generalized linear models. Research indicates that under certain conditions, particularly with sufficient sample size and specific model configurations, early and late fusion can be mathematically equivalent [2]. However, early fusion may fail to capture complex relationships when nonlinear feature-label associations exist across modalities [2].
Comparative Analysis of Fusion Strategies
Data-Level (Early) Fusion
Data-level fusion involves the direct combination of raw data from multiple sources into a single, coherent dataset before any significant feature extraction or modeling occurs [26]. This approach is also commonly referred to as early fusion or input fusion [25].
Mechanism and Workflow: The process typically begins with the temporal and spatial registration of data from different sources to ensure alignment. The core step is the concatenation or aggregation of these aligned raw data inputs into a unified structure. This combined dataset then serves as the input for a single, monolithic model which handles both the learning of cross-modal relationships and the final prediction [27].
Advantages: The primary strength of data-level fusion lies in its potential to preserve all information present in the original data. By combining data at the rawest level, the model has direct access to the complete dataset, which, in theory, allows it to learn complex, low-level interactions between modalities that might be lost during pre-processing or feature extraction [26]. The workflow is also relatively straightforward, requiring the training and maintenance of only one model.
Disadvantages and Challenges: A significant drawback is the curse of dimensionality; merging raw data can create very high-dimensional input vectors, which increases the risk of overfitting, especially with limited training samples [26]. This approach is also highly susceptible to data quality issues like noise and misalignment, as errors in one modality can directly corrupt the fused dataset. Furthermore, data-level fusion is inflexible, as adding a new data source typically requires rebuilding the entire dataset and retraining the model from scratch.
Feature-Level (Intermediate) Fusion
Feature-level fusion, or intermediate fusion, strikes a balance between the early and late approaches. Here, features are first extracted independently from each data source, and these feature vectors are then combined into a joint representation before being fed into a classifier [25].
Mechanism and Workflow: The process involves two key stages. First, modality-specific features are learned or engineered separately. In deep learning contexts, this is often done using dedicated neural network branches for each data type. Second, these feature sets are integrated through methods like concatenation, element-wise addition, or more sophisticated attention-based mechanisms that weight the importance of different features [25]. The fused feature vector is then processed by a final classification network.
Advantages: This strategy allows the model to learn rich inter-modal interactions at a meaningful abstraction level, without the noise and dimensionality problems of raw data fusion [25] [27]. It is more flexible than data-level fusion, as individual feature extractors can be updated independently. Hierarchical and attention-based fusion techniques within this category can model complex, non-linear relationships between features from different sources [25].
Disadvantages and Challenges: The main challenge is the increased architectural complexity, requiring careful design of the fusion layer (e.g., choosing between concatenation, attention, etc.) [25]. It also demands that all modalities are available during training to learn the cross-modal correlations, and performance can be sensitive to the quality and scale of the initial feature extraction.
Decision-Level (Late) Fusion
Decision-level fusion, known as late fusion, is a modular approach where separate models are trained for each data modality, and their individual predictions are aggregated to form a final decision [2] [26].
Mechanism and Workflow: In this strategy, a dedicated classifier is trained on each modality independently. At inference time, each model processes its respective input and outputs a prediction (e.g., a class probability or a regression value). These individual decisions are then combined using an aggregation function, which can be as simple as a weighted average or majority voting, or a more complex meta-classifier that learns to weigh the models based on their reliability [2] [27].
Advantages: Late fusion offers superior modularity and flexibility. New data sources can be incorporated by simply adding a new model and including its output in the aggregation function, without retraining the existing system [26]. It also avoids the dimensionality issues of early fusion and allows for the use of highly specialized, optimized models for each data type. This approach is naturally robust to missing modalities, as the system can still function, albeit with potentially reduced accuracy, if one model's input is unavailable.
Disadvantages and Challenges: The most significant limitation is the potential loss of inter-modality information. Because models are trained in isolation, crucial cross-modal dependencies (e.g., the correlation between a specific genetic marker and a visual symptom) may not be captured [26]. Training and maintaining multiple models can also be computationally expensive and logistically complex.
Diagram 1: Architectural comparison of data fusion strategies, showing the stage at which fusion occurs in each paradigm.
Table 2: Strategic Comparison of Data Fusion Levels
| Characteristic | Data-Level (Early) Fusion | Feature-Level (Intermediate) Fusion | Decision-Level (Late) Fusion |
|---|---|---|---|
| Fusion Stage | Input level | Feature level | Decision level |
| Information Preservation | High (raw data) | Moderate (processed features) | Low (model outputs) |
| Inter-Modality Interaction | Direct and potentially comprehensive | Learned and hierarchical | Limited or none |
| Dimensionality | Very High | Moderate | Low |
| Flexibility to New Modalities | Low (requires full retraining) | Moderate (may require fusion layer adjustment) | High (add new model) |
| Robustness to Missing Data | Low | Low | High |
| Computational Complexity | Single, potentially complex model | Multiple feature extractors + fusion network | Multiple independent models |
| Implementation Complexity | Low to Moderate | High | Moderate |
Experimental Evidence and Performance Comparison
Key Experimental Protocols
Empirical evaluations across diverse fields consistently demonstrate the performance advantages of data fusion over single-modality approaches. A rigorous experimental protocol for comparing fusion strategies typically involves several key stages.
First, data acquisition and preprocessing must be standardized. For instance, in a study on genomic and phenotypic selection (GPS), genomic data (e.g., SNP markers) and phenotypic data (e.g., crop yield, plant height) were collected from multiple crop species including maize, soybean, rice, and wheat [3]. Similarly, a study on Normal Tissue Complication Probability (NTCP) modeling for osteoradionecrosis used clinical variables (demographic and treatment data) and mandibular radiation dose distribution volumes (dosiomics) [28].
The core of the protocol involves model training and validation. Researchers typically implement all three fusion strategies using consistent underlying algorithms (e.g., Lasso, Random Forests, SVM, or deep learning architectures like 3D DenseNet) to ensure fair comparison [3] [28]. Performance is evaluated through rigorous cross-validation, measuring metrics such as accuracy, area under the curve (AUC), calibration, and robustness to varying sample sizes or data quality.
Quantitative Performance Comparison
Recent experimental findings provide compelling evidence for the superior predictive accuracy of data fusion compared to single-source techniques.
Table 3: Experimental Performance Comparison Across Domains
| Application Domain | Superior Fusion Strategy | Performance Gain Over Best Single-Source Model | Key Experimental Findings |
|---|---|---|---|
| Genomic & Phenotypic Selection (GPS) in Crops [3] | Data-Level (Early) Fusion | 53.4% improvement over best genomic model; 18.7% over best phenotypic model | Lasso_D model showed high accuracy with sample sizes as small as 200; robust to SNP density variations |
| Mandibular Osteoradionecrosis NTCP Modelling [28] | Decision-Level (Late) Fusion | Superior discrimination and calibration vs. single-modality models | Late fusion outperformed joint fusion; no significant discrimination difference between strategies |
| Theoretical Analysis (Generalized Linear Models) [2] | Context-Dependent | Critical sample size threshold for performance reversal | Early fusion outperforms with small samples; late fusion excels beyond critical sample size |
In the agricultural study, the data fusion framework (GPS) was tested with an extensive suite of models including GBLUP, BayesB, Lasso, RF, SVM, XGBoost, LightGBM, and DNNGP. The data-level fusion strategy consistently achieved the highest accuracy, with the top-performing model (Lasso_D) demonstrating exceptional robustness even with limited sample sizes and varying data conditions [3].
In the medical domain, a comparative study on osteoradionecrosis NTCP modelling found that while late fusion exhibited superior discrimination and calibration, joint (intermediate) fusion achieved a more balanced distribution of predicted probabilities [28]. This highlights that the optimal strategy may depend on the specific performance metric prioritized by researchers.
A crucial theoretical insight explains these contextual results: the existence of a critical sample size threshold at which the performance dominance of early and late fusion reverses [2]. This provides a mathematical foundation for why different strategies excel under different data conditions, moving the field beyond trial-and-error selection.
The Scientist's Toolkit: Research Reagent Solutions
Implementing effective data fusion requires both conceptual understanding and practical tools. The following table outlines essential "research reagents" - key algorithms, software, and data management solutions - that form the foundation for data fusion experiments in scientific research.
Table 4: Essential Research Reagent Solutions for Data Fusion Experiments
| Tool Category | Specific Examples | Function in Data Fusion Research |
|---|---|---|
| Machine Learning Algorithms | Lasso, Random Forests (RF), Support Vector Machines (SVM) [3] | Base models for feature selection and classification in fusion frameworks |
| Deep Learning Architectures | 3D DenseNet-40 [28], DNNGP [3] | Processing complex image data (e.g., dose distributions); genomic prediction |
| Ensemble Methods | Weighted Averaging, Stacking, Majority Voting [26] | Aggregating predictions from multiple models in late fusion |
| Fusion-Specific AI | Diag2Diag AI [29] [30] | Generating synthetic sensor data to recover missing information |
| Data Platforms & Ecosystems | Fusion Energy Data Ecosystem and Repository (FEDER) [31] | Unified platform for sharing standardized fusion data across institutions |
| Feature Extraction Techniques | Attention Mechanisms [25], Hierarchical Fusion | Identifying and weighting important features across modalities |
Specialized tools like the Diag2Diag AI demonstrate advanced fusion applications, capable of generating synthetic sensor data to compensate for missing diagnostic information in complex systems like fusion energy reactors [29] [30]. This approach has particular relevance for pharmaceutical research where complete multimodal datasets are often difficult to obtain.
Similarly, community-driven initiatives like the Fusion Energy Data Ecosystem and Repository (FEDER) highlight the growing importance of standardized data platforms for accelerating research through shared resources and reproducible workflows [31]. While developed for fusion energy, this concept directly translates to drug development where multi-institutional collaboration is common.
The comparative analysis of data-level, feature-level, and decision-level fusion strategies reveals a consistent theme: multimodal data fusion generally enhances predictive accuracy compared to single-technique approaches, but the optimal strategy is highly context-dependent. Experimental evidence from diverse fields including agriculture, medical physics, and energy research demonstrates performance improvements ranging from approximately 19% to over 53% when employing an appropriate fusion strategy compared to the best single-source model [3] [28].
The selection of an appropriate fusion strategy depends on multiple factors, including data characteristics (volume, quality, modality relationships), computational resources, and project requirements for flexibility and robustness. Data-level fusion often excels with smaller datasets and strong inter-modal dependencies [2] [3]. Feature-level fusion offers a balanced approach for capturing complex interactions without the dimensionality curse of raw data fusion [25]. Decision-level fusion provides maximum flexibility and robustness, particularly valuable when dealing with missing data or frequently added new modalities [28] [26].
For researchers and drug development professionals, these findings underscore the importance of systematically evaluating multiple fusion strategies rather than relying on a default approach. The emerging paradigm of fusion method selection, guided by theoretical principles like critical sample size thresholds and data relationship classifications, promises to enhance research efficiency and predictive accuracy in the increasingly multimodal landscape of scientific inquiry [2] [24].
In the fields of genomics and drug development, a significant challenge persists: connecting an organism's genetic blueprint (genotype) to its observable characteristics (phenotype). Traditional approaches that rely on a single data type often yield incomplete insights, especially for complex traits influenced by multiple genetic and environmental factors [32]. Data integration is increasingly becoming an essential tool to cope with the ever-increasing amount of data, to cross-validate noisy datasets, and to gain broad interdisciplinary views of large genomic and proteomic datasets [32].
This guide objectively compares the performance of a data fusion strategy against single-technique research, framing the discussion within the broader thesis that integrating disparate data sources provides superior predictive accuracy than any individual dataset alone. The overarching goals of data integration are to obtain more precision, better accuracy, and greater statistical power [32]. By synthesizing methodologies and evidence from current research, we provide a framework for researchers and scientists to construct effective fusion pipelines, thereby unlocking more reliable and actionable biological insights.
Data Fusion Strategies: A Comparative Framework
Data fusion methods can be systematically classified based on the stage at which integration occurs. Understanding this hierarchy is crucial for selecting the appropriate architectural strategy for a genomic-phenotypic pipeline [2].
Table: Classification of Data Fusion Strategies
| Fusion Type | Alternative Names | Stage of Integration | Key Characteristics |
|---|---|---|---|
| Early Fusion | Data-Level Fusion | Input Stage | Raw or pre-processed data from multiple sources are concatenated into a single feature set before model input. |
| Intermediate Fusion | Feature-Level Fusion | Processing Stage | Features are extracted from each data source independently first, then combined into a joint feature space. |
| Late Fusion | Decision-Level Fusion | Output Stage | Separate models are trained on each data type, and their predictions are aggregated for a final decision. |
The following diagram illustrates the workflow and data flow for these three primary fusion strategies:
Performance Comparison: Fusion vs. Single-Technique Approaches
Recent large-scale studies provide compelling experimental data on the performance benefits of data fusion. A 2025 study introduced the GPS (Genomic and Phenotypic Selection) framework, rigorously testing it on datasets from four crop species: maize, soybean, rice, and wheat [3]. The study compared three fusion strategies—data fusion (early), feature fusion (intermediate), and result fusion (late)—against standalone Genomic Selection (GS) and Phenotypic Selection (PS) models using a suite of statistical and machine learning models [3].
Table: Predictive Performance Comparison of Fusion vs. Single-Technique Models
| Model Type | Specific Model | Key Performance Finding | Comparative Advantage |
|---|---|---|---|
| Best Data Fusion Model | Lasso_D | Improved accuracy by 53.4% vs. best GS model and 18.7% vs. best PS model [3]. | Highest overall accuracy, robust to sample size and SNP density variations. |
| Best Genomic Selection (GS) Model | LightGBM | Used as baseline for GS accuracy [3]. | Effective for purely genetic prediction, but outperformed by fusion. |
| Best Phenotypic Selection (PS) Model | Lasso | Used as baseline for PS accuracy [3]. | Effective for trait correlation, but outperformed by fusion. |
| Feature Fusion | Various | Lower accuracy than data fusion in GPS study [3]. | Provides a middle ground, fusing extracted features. |
| Result Fusion | Various | Lower accuracy than data fusion in GPS study [3]. | Leverages model diversity, but decision aggregation can dilute performance. |
Beyond raw accuracy, the fusion model Lasso_D demonstrated exceptional robustness and transferability. It maintained high predictive accuracy with sample sizes as small as 200 and was resilient to variations in single-nucleotide polymorphism (SNP) density [3]. Furthermore, in cross-environmental prediction scenarios—a critical test for real-world application—the model showed only a 0.3% reduction in accuracy compared to predictions generated using data from the same environment [3].
Experimental Protocols: Methodologies for Validated Comparisons
To ensure the reproducibility of fusion pipeline results, this section outlines the detailed experimental protocols and methodologies employed in the key studies cited.
The GPS Framework Experimental Protocol
The following workflow was used to generate the comparative performance data in the GPS study [3]:
Key Experimental Details:
- Datasets: Large-scale genomic and phenotypic data from four crop species (maize, soybean, rice, wheat) were used to ensure broad applicability and robustness [3].
- Model Suite: The study employed a diverse set of models for comprehensive comparison, including statistical approaches (GBLUP, BayesB), traditional machine learning (Lasso, RF, SVM, XGBoost, LightGBM), and deep learning (DNNGP) [3].
- Validation: Rigorous cross-validation techniques were applied to assess model performance, robustness to sample size and SNP density, and transferability across different environments [3].
Theoretical Selection Protocol
Beyond empirical testing, a 2025 mathematical analysis established a paradigm for selecting the optimal fusion method prior to task execution [2]. This protocol helps researchers avoid computationally expensive trial-and-error approaches.
Key Theoretical Findings and Selection Criteria:
- Equivalence Conditions: The study derived the specific mathematical conditions under which early and late fusion methods produce equivalent results within the framework of generalized linear models [2].
- Failure Conditions for Early Fusion: The analysis identified scenarios where early fusion is likely to underperform, particularly in the presence of nonlinear feature-label relationships [2].
- Critical Sample Size Threshold: A key contribution was the proposal of a critical sample size threshold at which the performance dominance of early and late fusion models reverses. This provides a quantitative guide for method selection based on dataset size [2].
- Selection Paradigm: Based on these derivations, the researchers introduced a fusion method selection paradigm that uses factors like sample size, feature quantity, and modality number to recommend the most appropriate fusion method before model training [2].
The Scientist's Toolkit: Essential Reagents and Computational Solutions
Building an effective genomic-phenotypic fusion pipeline requires both physical research reagents and computational tools. The following table details key components and their functions.
Table: Essential Research Reagents and Computational Solutions
| Category | Item | Specific Function in Pipeline |
|---|---|---|
| Genomic Data Generation | Next-Generation Sequencing (NGS) | Provides high-throughput genomic data, including SNPs and structural variants, forming one core data modality [33]. |
| Phenotypic Data Acquisition | High-Throughput Phenotyping Systems (e.g., drones, sensors) | Automated collection of large-scale phenotypic data (e.g., plant size, architecture) from field or lab conditions [33]. |
| Phenotypic Data Acquisition | ChronoRoot 2.0 Platform | An open-source tool that uses AI to track and analyze multiple plant structures, such as root architecture, over time [33]. |
| Intermediate Data Layers | Multi-Omics Technologies (Transcriptomics, Proteomics, Metabolomics) | Provides intermediate molecular layers that help bridge the gap between genotype and phenotype for a more complete picture [33]. |
| Computational & AI Tools | PhenoAssistant | Employs Large Language Models (LLMs) to orchestrate phenotype extraction, visualization, and model training from complex datasets [33]. |
| Computational & AI Tools | Genomic Language Models | Treat DNA sequences as a language to predict the functional impact of genetic variants and detect regulatory elements [33]. |
| Computational & AI Tools | Explainable AI (XAI) Tools | Critical for interpreting complex fusion models, providing biological insights, and building trust in predictions by explaining the model's reasoning [33]. |
The empirical evidence and methodological frameworks presented in this guide consistently support the thesis that data fusion strategies significantly enhance predictive accuracy compared to single-technique genomic or phenotypic approaches. The integration of genomic and phenotypic data addresses the inherent complexity of biological systems, where observable traits are rarely the product of a single genetic factor.
The future of genomic-phenotypic fusion lies in more seamless and automated pipelines. Key emerging trends include the increased use of Generative AI for in-silico experimentation and data augmentation, a stronger focus on modeling temporal and spatial dynamics of traits, and the development of closed-loop systems that integrate AI-based prediction with genome editing (e.g., CRISPR) to rapidly test and validate biological hypotheses [33]. As these technologies mature, the fusion of genomic and phenotypic data will undoubtedly become a standard, indispensable practice in basic biological research and applied drug development.
Cancer survival prediction remains a pivotal challenge in precision oncology, directly influencing therapeutic decisions and patient management. Traditional prognostic models, often reliant on single data types like clinical variables or genomic markers, frequently fail to capture the complex molecular heterogeneity that drives patient-specific outcomes. The integration of multiple omics technologies—known as multi-omics fusion—has emerged as a transformative approach that leverages complementary biological information to achieve more accurate and robust predictions. This case study examines the paradigm of data fusion predictive accuracy versus single-technique research through the lens of cancer survival prediction, demonstrating how integrated analytical frameworks consistently outperform unimodal approaches across multiple cancer types and methodological implementations.
Recent technological advancements have generated vast amounts of molecular data through high-throughput sequencing and other molecular assays, creating unprecedented opportunities for comprehensive cancer profiling. Large-scale initiatives such as The Cancer Genome Atlas (TCGA) provide publicly available datasets that integrate clinical, omics, and histopathology imaging data for thousands of cancer patients [34] [35]. Concurrently, artificial intelligence (AI) and machine learning (ML) have empowered the development of sophisticated fusion models capable of managing the high dimensionality and heterogeneity of these multimodal datasets [35] [8]. This convergence of data availability and computational innovation has positioned multi-omics fusion as a cornerstone of next-generation cancer prognostic tools.
Performance Comparison: Multi-omics Fusion vs. Single-Modality Approaches
Quantitative evidence from recent studies consistently demonstrates the superior predictive performance of multimodal data integration compared to single-modality approaches. The comparative analysis reveals that fusion models achieve significant improvements in key prognostic metrics across diverse cancer types.
Table 1: Performance Comparison of Fusion Strategies vs. Single Modalities in Cancer Survival Prediction
| Cancer Type | Model/Framework | Data Modalities | Fusion Strategy | Performance (C-Index) | Superiority vs. Single Modality |
|---|---|---|---|---|---|
| Breast Cancer | Multimodal Deep Learning [34] | Clinical, Somatic SNV, RNA-seq, CNV, miRNA, Histopathology | Late Fusion | Highest test-set concordance | Consistently outperformed early fusion and unimodal approaches |
| Pan-Cancer | AZ-AI Multimodal Pipeline [8] | Transcripts, Proteins, Metabolites, Clinical factors | Late Fusion | Improved accuracy & robustness | Outperformed single-modality approaches in TCGA lung, breast, and pan-cancer datasets |
| Breast Cancer | Adaptive Multi-omics Integration [36] | Genomics, Transcriptomics, Epigenomics | Genetic Programming | C-index: 67.94 (test set) | Demonstrated potential of integrated approach over traditional single-omics |
| Multiple Cancers | M3Surv [37] | Multi-slide pathology, Multi-omics | Hybrid Fusion | Average 2.2% C-index improvement | Outperformed state-of-the-art methods and showed stability with missing modalities |
| Liver & Breast Cancer | DeepProg [36] | Multi-omics | Unspecified | C-index: 0.68-0.80 | Robustly predicted survival subtypes across cancer types |
The performance advantage of multi-omics fusion extends beyond single cancer types, demonstrating generalizability across malignancies. A systematic review of 196 studies on machine learning for cancer survival analysis found that "improved predictive performance was seen from the use of ML in almost all cancer types," with multi-task and deep learning methods yielding superior performance [38]. This pattern establishes a consistent trend wherein models leveraging complementary data sources achieve enhanced prognostic capability compared to their single-modality counterparts.
Table 2: Advantages and Limitations of Multi-omics Fusion Strategies
| Fusion Strategy | Key Advantages | Limitations | Representative Performance |
|---|---|---|---|
| Early Fusion (Data-level) | Enables learning of joint representations; Useful for temporally/spatially aligned modalities | Prone to overfitting with high-dimensional data; Poor gradient flow; Discards platform heterogeneity | Lower performance compared to late fusion in high-dimensional settings [34] [35] |
| Late Fusion (Decision-level) | Enhanced resistance to overfitting; Handles data heterogeneity; Allows modality-specific specialization | May miss cross-modal interactions; Difficulties identifying inter-omics relationships | Consistently outperformed early fusion in multiple studies [34] [8] |
| Hybrid Fusion | Combines strengths of both approaches; Enhances biological relevance | Increased complexity; More challenging to implement and train | M3Surv achieved 2.2% average C-index improvement [37] |
| Intermediate Integration | Balances cross-modal learning with modality-specific processing | Still vulnerable to dimensionality challenges | Varies by implementation; Generally outperforms early fusion [35] |
Experimental Protocols and Methodologies
Data Acquisition and Preprocessing Frameworks
Robust multi-omics fusion begins with systematic data acquisition and preprocessing. Recent studies have established standardized pipelines for handling diverse molecular data types from sources such as TCGA. A characteristic framework is exemplified by the breast cancer survival prediction study that integrated clinical variables, somatic mutations, RNA expression, copy number variation, miRNA expression, and histopathology images from TCGA [34]. Their preprocessing protocol employed distinct filtering strategies for each modality: somatic SNV data were processed into a binary sample-by-gene matrix with a 1% mutation frequency threshold; RNA-seq data (in FPKM units) were restricted to cancer-related genes in the CGN MSigDB gene set; CNV data were normalized to a range of -2 to 2; and miRNA expression profiles were filtered to retain only miRNAs altered in at least 10% of the cohort [34]. This modality-specific preprocessing ensures optimal feature relevance while managing dimensionality.
The MLOmics database provides another comprehensive preprocessing pipeline, offering three feature versions (Original, Aligned, and Top) tailored to various machine learning tasks [39]. Their Top version employs multi-class ANOVA to identify genes with significant variance across cancer types, followed by Benjamini-Hochberg correction to control the false discovery rate, and finally z-score normalization [39]. Such standardized preprocessing addresses the critical challenge of platform compatibility and data heterogeneity that often impedes multi-omics integration.
Fusion Methodologies and Model Architectures
The core innovation in multi-omics survival prediction lies in the fusion methodologies that integrate disparate data types. The AZ-AI multimodal pipeline exemplifies a versatile approach that compares early, intermediate, and late fusion strategies [8]. In their implementation, late fusion—processing each modality independently through dedicated networks and combining outputs at the decision level—consistently outperformed other strategies, particularly in settings with high-dimensional feature spaces (10³-10⁵ features) and limited samples (10-10³ patients) [8]. This modular approach allows each sub-network to specialize in a particular data type, improving interpretability and model robustness when modalities vary in data quality or availability.
Another advanced architecture, M3Surv, addresses the critical challenge of missing modalities through a hypergraph-based learning approach and prototype-based memory bank [37]. Their framework captures both intra-slide higher-order cellular structures and inter-slide relationships from multiple pathology slides, then integrates multi-omics data using interactive cross-attention mechanisms [37]. The memory bank learns and stores representative pathology-omics feature prototypes during training, enabling robust imputation when modalities are entirely missing at inference—a common scenario in clinical practice. This innovation demonstrates how sophisticated fusion architectures can overcome real-world data incompleteness while maintaining predictive performance.
Validation Frameworks and Evaluation Metrics
Rigorous validation is essential for accurate performance assessment in multi-omics fusion studies. The leading approaches employ comprehensive frameworks based on cross-validation with fixed held-out test sets to ensure unbiased evaluation of out-of-sample performance [34] [8]. For instance, one breast cancer study defined a fixed test set comprising 175 patients (20% of the smallest unimodal dataset), with the remaining samples partitioned into five stratified folds for cross-validation while maintaining balanced outcome representation [34]. This methodology prevents overoptimistic performance estimates and ensures reliable generalizability assessment.
The concordance index (C-index) serves as the primary evaluation metric for survival prediction, measuring the proportion of patient pairs correctly ordered by the model [36]. Additional metrics include precision, recall, and F1-score for classification tasks, plus normalized mutual information (NMI) and adjusted rand index (ARI) for clustering evaluation [39]. The systematic implementation of these robust validation protocols and standardized metrics enables meaningful comparison across different fusion strategies and establishes credible performance benchmarks for the field.
Visualization of Multi-omics Fusion Workflows
Generalized Multi-omics Fusion Pipeline
The following diagram illustrates a comprehensive workflow for multi-omics data fusion in cancer survival prediction, integrating elements from multiple studies [34] [8] [37]:
Multi-omics Fusion Strategy Comparison
This diagram illustrates the architectural differences between early, intermediate, and late fusion strategies:
Successful implementation of multi-omics fusion requires specialized computational tools, datasets, and methodologies. The following table catalogs essential "research reagent solutions" that form the foundation of this field.
Table 3: Essential Research Reagents and Resources for Multi-omics Fusion
| Resource Category | Specific Tool/Database | Key Function | Application Context |
|---|---|---|---|
| Public Data Repositories | The Cancer Genome Atlas (TCGA) | Provides comprehensive molecular and clinical data across cancer types | Primary data source for model development and validation [34] [35] |
| Public Data Repositories | The Cancer Imaging Archive (TCIA) | Stores medical images correlated with genomic and clinical data | Facilitates radiogenomic studies and imaging-omics integration [35] |
| Public Data Repositories | MLOmics Database | Preprocessed, ML-ready multi-omics data with standardized features | Reduces preprocessing burden; enables fair model comparisons [39] |
| Computational Frameworks | AZ-AI Multimodal Pipeline | Versatile Python library for multimodal feature integration and survival prediction | Supports comparison of fusion strategies and feature reduction methods [8] |
| Computational Frameworks | M3Surv Framework | Hypergraph-based fusion of multi-slide pathology and multi-omics data | Addresses missing modality challenge via prototype-based memory bank [37] |
| Computational Frameworks | Adaptive Multi-omics Integration with Genetic Programming | Evolves optimal integration of omics data using evolutionary algorithms | Optimizes feature selection and integration for biomarker discovery [36] |
| Bioinformatics Tools | STRING Database | Protein-protein interaction networks and functional enrichment analysis | Biological validation and interpretation of molecular features [39] |
| Bioinformatics Tools | KEGG Pathway Database | Reference knowledge base for pathway mapping and functional annotation | Contextualizes findings within established biological pathways [39] |
| Evaluation Metrics | Concordance Index (C-index) | Measures model discrimination ability for survival predictions | Standardized performance assessment across studies [8] [36] |
| Evaluation Metrics | Normalized Mutual Information (NMI) | Evaluates clustering quality in subtype discovery | Assesses agreement between computational results and known biology [39] |
The evidence presented in this case study substantiates the thesis that data fusion predictive accuracy significantly surpasses single-technique research in cancer survival prediction. Multi-omics integration consistently demonstrates superior performance across multiple cancer types, with late fusion strategies emerging as particularly effective for high-dimensional omics data. The methodological advances in fusion architectures, coupled with robust validation frameworks and specialized computational tools, have established a new paradigm in prognostic modeling.
The translational impact of these approaches extends beyond improved prediction metrics. Multi-omics fusion enables more precise patient stratification, identification of novel biomarkers, and deeper insights into the complex molecular mechanisms driving cancer progression [36] [40]. Furthermore, frameworks capable of handling missing modalities bridge the gap between research ideals and clinical reality, where complete multi-omics profiling is often impractical [37]. As these technologies mature and standardization improves, multi-omics fusion is poised to become an indispensable component of precision oncology, ultimately contributing to more personalized treatment strategies and improved patient outcomes.
Genomic selection (GS) has revolutionized plant breeding by enabling the prediction of breeding values using genome-wide markers, thereby accelerating genetic gain [41] [42]. However, the predictive accuracy of traditional GS models is often constrained by the complex architecture of quantitative traits and the limited information captured by genomic markers alone [43]. In response, data fusion strategies have emerged as a powerful approach to enhance prediction performance by integrating complementary data sources [3] [43]. This case study provides a comparative analysis of leading genomic prediction methodologies, with a specific focus on evaluating the performance of data fusion techniques against conventional single-technique approaches. We examine experimental data from diverse crop species to offer plant breeders and researchers evidence-based guidance for implementing these advanced predictive frameworks.
Comparative Performance of Genomic Prediction Methods
Performance Benchmarking Across Models and Crops
Table 1: Comparative performance of genomic prediction methods across crop species
| Method Category | Specific Method | Crop | Trait | Prediction Accuracy | Key Advantage |
|---|---|---|---|---|---|
| Data Fusion | Lasso_D (GPS Framework) | Maize, Soybean, Rice, Wheat | Complex traits | 53.4% improvement over best GS model (LightGBM) [3] | Highest accuracy strategy; robust to sample size & SNP density [3] |
| Deep Learning | WheatGP (CNN-LSTM) | Wheat | Yield | 0.73 [44] | Captures additive and epistatic effects [44] |
| Deep Learning | DNNGP | Wheat | Grain length | 0.766 [44] | Multi-layered hierarchical feature learning [44] |
| Deep Learning | SoyDNGP | Soybean | Hundred-seed weight | 0.836 [44] | Species-optimized architecture [44] |
| Machine Learning | LightGBM | Multiple crops | Various | Baseline for GS [3] | Computational efficiency [45] |
| Machine Learning | Lasso | Multiple crops | Various | Baseline for PS [3] | Feature selection capability [46] |
| Bayesian | Bayes B | Soybean, Rice, Maize | Traits with varying heritability | Superior for traits with major-effect loci [46] | Variable selection for markers with large effects [46] |
| Multi-omics Integration | Model-based Fusion | Maize, Rice | Complex traits | Consistent improvement over genomic-only [43] | Captures non-additive, nonlinear interactions [43] |
Key Performance Insights
The comparative data reveal several critical patterns. First, data fusion strategies, particularly the GPS framework with Lasso_D, demonstrate substantial improvements over single-technique approaches, outperforming the best genomic selection model by 53.4% and the best phenotypic selection model by 18.7% [3]. Second, species-specific deep learning models like WheatGP and SoyDNGP achieve remarkably high accuracy for particular crop-trait combinations, with SoyDNGP reaching 0.836 for hundred-seed weight in soybean [44]. Third, the performance of traditional methods varies significantly by context; for instance, Bayes B shows particular strength for traits with known major-effect loci [46].
Experimental Protocols and Methodologies
Data Fusion Framework (GPS) Experimental Protocol
Experimental Objective: To evaluate three data fusion strategies (data fusion, feature fusion, and result fusion) for integrating genomic and phenotypic data to improve prediction accuracy of complex traits in plants [3] [47].
Materials and Methods:
- Plant Materials: Large datasets from four crop species: maize, soybean, rice, and wheat [3]
- Genotypic Data: Single-nucleotide polymorphism (SNP) markers with varying densities to test model robustness [3]
- Phenotypic Data: Multiple agronomic traits with varying heritability and genetic architecture [3]
- Comparison Models: Statistical approaches (GBLUP, BayesB), machine learning models (Lasso, RF, SVM, XGBoost, LightGBM), deep learning method (DNNGP), and a recent phenotype-assisted prediction model (MAK) [3]
- Fusion Strategies:
- Data Fusion: Early integration of raw genomic and phenotypic data before model application
- Feature Fusion: Intermediate integration of extracted features from separate genomic and phenotypic models
- Result Fusion: Late integration through weighted averaging of predictions from separate genomic and phenotypic models [3]
- Evaluation Metrics: Prediction accuracy measured as correlation between predicted and observed values, robustness to sample size and SNP density, and transferability across environments [3]
WheatGP Deep Learning Architecture Protocol
Experimental Objective: To develop a genomic prediction method for wheat that captures both additive genetic effects and epistatic genetic effects through a combined CNN-LSTM architecture [44].
Materials and Methods:
- Plant Materials: Wheat599 dataset (599 varieties, 1279 markers) and wheat2000 dataset (2000 Iranian bread wheat varieties, 33,709 markers) [44]
- Traits Evaluated: Grain yield and six agronomic traits: grain length, grain width, grain hardness, thousand-kernel weight, test weight, and grain protein [44]
- Architecture Components:
- CNN Module: Three convolutional layers with ReLU activation to capture short-range dependencies within genomic sequences
- LSTM Module: Gating mechanism to retain long-distance dependency relationships between gene loci
- Prediction Module: Fully connected layer to map extracted features to phenotype predictions [44]
- Training Specifications: Bayesian-based hyperparameter optimization within Optuna framework for batch size, weight decay, and learning rate [44]
- Comparison Models: Benchmarking against rrBLUP, XGBoost, SVR, and DNNGP [44]
- Evaluation Metric: Pearson correlation coefficient (PCC) between predicted and observed values using 10-fold cross-validation [44]
Multi-omics Integration Protocol
Experimental Objective: To assess 24 integration strategies combining three omics layers (genomics, transcriptomics, and metabolomics) for improving genomic prediction of complex traits [43].
Materials and Methods:
- Datasets: Three real-world datasets from maize and rice with variations in population size, trait complexity, and omics dimensionality:
- Maize282 (279 lines, 22 traits, 50,878 markers, 18,635 metabolomic features, 17,479 transcriptomic features)
- Maize368 (368 lines, 20 traits, 100,000 markers, 748 metabolomic features, 28,769 transcriptomic features)
- Rice210 (210 lines, 4 traits, 1,619 markers, 1,000 metabolomic features, 24,994 transcriptomic features) [43]
- Integration Approaches:
- Early Data Fusion: Concatenation of raw omics data
- Model-Based Integration: Advanced statistical and machine learning methods capable of capturing non-additive, nonlinear, and hierarchical interactions across omics layers [43]
- Evaluation: Predictive accuracy comparison against genomic-only models using standardized cross-validation procedures [43]
Visualizing Methodologies and Workflows
Data Fusion Strategies Workflow
WheatGP Neural Network Architecture
Table 2: Key research reagents and computational resources for genomic prediction studies
| Resource Category | Specific Resource | Application in Genomic Prediction | Key Features/Benefits |
|---|---|---|---|
| Benchmarking Platforms | EasyGeSe [45] | Standardized benchmarking of genomic prediction methods | Curated datasets from multiple species; standardized evaluation procedures [45] |
| Statistical Software | BGLR Statistical Package [42] | Bayesian regression models for genomic prediction | Implements various prior distributions for marker effects [42] |
| Machine Learning Libraries | XGBoost, LightGBM [48] [45] | Gradient boosting for genomic prediction | Computational efficiency; handles high-dimensional data [45] |
| Deep Learning Frameworks | TensorFlow, PyTorch [44] | Implementing CNN, LSTM architectures | Flexible deep learning model development [44] |
| Hyperparameter Optimization | Optuna Framework [44] | Automated hyperparameter tuning for machine/deep learning | Bayesian optimization; efficient parameter space exploration [44] |
| Genomic Datasets | Wheat599, Wheat2000 [44] | Training and validation of prediction models | Publicly available standardized datasets [44] |
| Multi-omics Datasets | Maize282, Maize368, Rice210 [43] | Multi-omics integration studies | Combined genomic, transcriptomic, and metabolomic data [43] |
| Model Interpretation | SHAP (SHapley Additive exPlanations) [44] | Interpreting complex model predictions | Feature importance analysis for genomic prediction [44] |
Discussion and Future Perspectives
The evidence from comparative studies indicates that data fusion approaches, particularly the GPS framework, represent a significant advancement in genomic prediction methodology. The superior performance of Lasso_D within this framework (53.4% improvement over the best GS model) demonstrates the value of integrating complementary data sources rather than relying on single-technique approaches [3]. This aligns with findings from multi-omics integration studies, where model-based fusion strategies consistently outperformed genomic-only models for complex traits [43].
The robustness of data fusion methods to variations in sample size and SNP density addresses critical limitations in plant breeding applications where these parameters are often constrained by practical and economic considerations [3]. Furthermore, the demonstrated transferability across environments with minimal accuracy reduction (0.3%) highlights the potential for applying these models in diverse breeding contexts [3].
Future research directions should focus on several key areas. First, developing more sophisticated integration frameworks for combining heterogeneous data types, including hyperspectral imaging, sensor data, and additional omics layers [49]. Second, creating more interpretable models that not only predict accurately but also provide biological insights into trait architecture, potentially through approaches like SHAP analysis [44]. Third, addressing the computational challenges associated with complex models through optimization and efficient implementation [45]. Finally, expanding the accessibility of these advanced methods to breeding programs through user-friendly tools and standardized benchmarking platforms like EasyGeSe [45].
As genomic selection continues to evolve, the integration of data fusion strategies with emerging artificial intelligence techniques will likely play a pivotal role in achieving the predictive accuracy required for Breeding 4.0, where the optimal combination of alleles for complex traits can be precisely predicted and efficiently incorporated into crop varieties [42].
Navigating the Challenges: A Framework for Optimizing Fusion Performance
In the pursuit of robust predictive models for drug development, researchers consistently encounter three interconnected pitfalls: data heterogeneity, high dimensionality, and overfitting. These challenges manifest with particular severity in pharmaceutical research, where the integration of diverse data modalities—from genomic sequences to clinical outcomes—has become standard practice. Data heterogeneity arises from combining information across multiple studies, platforms, and populations, introducing variability that can compromise analytical validity if not properly addressed [50]. Simultaneously, the curse of dimensionality presents fundamental obstacles when the number of features exponentially outpaces available samples, increasing computational demands while degrading model performance [51] [52]. These issues collectively predispose models to overfitting, where algorithms memorize noise and idiosyncrasies in training data rather than learning generalizable patterns.
The paradigm of data fusion, which integrates multiple data sources to create a unified predictive model, offers potential solutions yet introduces its own complexities. This guide objectively examines the performance of data fusion strategies against single-technique approaches, providing experimental data and methodologies to inform research decisions in pharmaceutical development.
Understanding Data Heterogeneity: Challenges and Identification
Defining Homogeneity and Heterogeneity in Research Data
In statistical terms, homogeneity describes datasets where elements share similar characteristics or statistical properties, while heterogeneity refers to datasets composed of dissimilar elements with varying statistical properties [53] [54]. In pharmaceutical research, this distinction extends beyond data composition to encompass variability in treatment effects, population responses, and experimental conditions across studies.
A homogeneous dataset exhibits consistent variance (homoscedasticity), where the variability of data points remains stable across the dataset [53]. Conversely, heterogeneous data demonstrates changing variances (heteroscedasticity), which violates key assumptions of many statistical models and can lead to biased standard errors and misleading inferences [53]. For drug development professionals, heterogeneity often manifests when pooling data from multiple clinical trials, where differences in study populations, protocols, or measurement techniques introduce systematic variations.
Statistical Tests for Identifying Heterogeneity
Researchers can employ several statistical methods to detect and quantify heterogeneity:
- Chi-Square Test for Homogeneity: Determines whether different populations follow the same distribution by comparing observed frequencies against expected frequencies across groups [54].
- Cochran's Q and I² Statistics: Meta-analysis measures that quantify the degree of heterogeneity across studies, with I² representing the percentage of total variability due to between-study differences rather than sampling error [50].
- Location Tests: Examine whether distributions share the same location parameter (e.g., mean or median) across different subgroups or studies [53].
The presence of heterogeneity necessitates specialized analytical approaches, as ignoring these variations can yield misleading conclusions and non-generalizable models.
The Curse of Dimensionality: Impacts and Mitigation Strategies
Fundamental Challenges in High-Dimensional Spaces
The "curse of dimensionality" describes phenomena that occur when analyzing data in high-dimensional spaces that do not appear in lower-dimensional settings [52]. In machine learning, this manifests through several interconnected problems:
- Data Sparsity: As dimensionality increases, the volume of space grows exponentially, causing available data points to become sparse and dissimilar [51] [52]. This sparsity undermines the ability to detect meaningful patterns.
- Distance Function Degradation: In high-dimensional spaces, Euclidean distances between points become increasingly similar, reducing the effectiveness of distance-based algorithms [52].
- Combinatorial Explosion: The number of possible feature combinations grows factorially, making exhaustive feature evaluation computationally prohibitive [52].
- Increased Overfitting Risk: Models with excessive parameters relative to training samples tend to memorize noise rather than learn generalizable relationships [55].
These challenges are particularly acute in genomics and pharmaceutical research, where datasets may contain thousands of genes or molecular features but only hundreds of patient samples [50] [52].
Dimensionality Reduction Techniques
Two primary approaches address high dimensionality: feature selection and feature extraction. Table 1 compares these approaches and their representative techniques.
Table 1: Dimensionality Reduction Techniques for High-Dimensional Data
| Approach | Description | Common Techniques | Best Use Cases |
|---|---|---|---|
| Feature Selection | Selects subset of most relevant features while discarding others | Low Variance Filter, High Correlation Filter, Forward Feature Construction, Backward Feature Elimination [56] | When interpretability is crucial; when original features must be retained |
| Feature Extraction | Transforms original features into new lower-dimensional space | PCA, LDA, t-SNE, UMAP, Autoencoders [51] [56] | When dealing with highly correlated features; when creating informative embeddings |
Principal Component Analysis (PCA) operates by identifying orthogonal principal components that maximize variance in the data [51] [56]. The algorithm follows these steps: (1) standardizing the original variables to zero mean and unit variance; (2) computing the covariance matrix to understand variable relationships; (3) calculating eigenvectors and eigenvalues of the covariance matrix; (4) sorting eigenvectors by decreasing eigenvalues; and (5) projecting the original data into the new subspace defined by the selected eigenvectors [56].
Linear Discriminant Analysis (LDA) differs from PCA by seeking feature combinations that maximize separation between classes rather than simply maximizing variance [56]. This supervised technique often achieves better performance for classification tasks in drug development contexts.
Manifold learning techniques such as t-Distributed Stochastic Neighbor Embedding (t-SNE) and Uniform Manifold Approximation and Projection (UMAP) excel at revealing intrinsic data structures in high-dimensional spaces [56]. These non-linear methods preserve local relationships between data points, making them particularly valuable for visualizing complex biological datasets.
Figure 1 illustrates the workflow for applying dimensionality reduction in pharmaceutical research:
Figure 1: Dimensionality Reduction Workflow for Pharmaceutical Data
Data Fusion vs. Single-Technique Approaches: Experimental Comparison
Methodologies for Comparative Evaluation
To objectively evaluate data fusion against single-technique approaches, we examine two experimental frameworks from recent literature:
Experiment 1: Drying Process Monitoring for Herbal Extracts This study compared Fourier Transform Near-Infrared Spectroscopy (FT-NIR) and Visible/Near-Infrared Hyperspectral Imaging (Vis/NIR-HSI) for monitoring critical quality attributes (CQAs) of JianWeiXiaoShi extract during pulsed vacuum drying [57]. The experimental protocol included:
- Sample Preparation: Herbal extracts were prepared with different excipients (maltodextrin and microcrystalline cellulose) and subjected to pulsed vacuum drying with sampling at predetermined intervals [57].
- Reference Analysis: Moisture content was measured using the loss-on-drying method, while marker compounds (narirutin and hesperidin) were quantified via high-performance liquid chromatography (HPLC) [57].
- Spectral Acquisition: FT-NIR spectra (800–2500 nm) and Vis/NIR-HSI images (400–1000 nm) were collected simultaneously throughout the drying process [57].
- Model Development: Partial least squares (PLS) regression models were built for individual techniques and for fused data using mid-level and high-level data fusion strategies [57].
Experiment 2: Pooled Genomic Analysis for Ovarian Cancer This analysis integrated clinical and microarray gene expression data from 2,970 ovarian cancer patients across 23 studies using 11 different gene expression measurement platforms [50]. The methodology included:
- Data Harmonization: Implementing rigorous curation procedures to standardize clinical annotations and normalize gene expression measurements across different platforms [50].
- Heterogeneity Assessment: Applying penalized Cox partial likelihood with adaptively weighted composite penalties to identify variables with homogeneous, heterogeneous, or null effects across studies [50].
- Model Implementation: Utilizing an iterative shooting algorithm to minimize the objective function with L1 and L1/L2 penalties on average and heterogeneous effects, respectively [50].
Quantitative Performance Comparison
Table 2 presents experimental results comparing prediction accuracy between single-technique approaches and data fusion strategies:
Table 2: Performance Comparison of Single-Technique vs. Data Fusion Approaches
| Application Domain | Single-Technique Approach | Performance Metric | Data Fusion Strategy | Performance Metric | Improvement |
|---|---|---|---|---|---|
| Herbal Extract Drying [57] | FT-NIR only | R² = 0.84-0.94 (CQAs) | FT-NIR + Vis/NIR-HSI (High-Level Fusion) | R² = 0.91-0.98 (CQAs) | 8.3-16.7% increase in R² |
| Herbal Extract Drying [57] | Vis/NIR-HSI only | R² = 0.89 (Color) | FT-NIR + Vis/NIR-HSI (High-Level Fusion) | R² > 0.95 (Color) | 6.7% increase in R² |
| Chemical Engineering Construction [6] | Standard Transformer | ~85.5% accuracy | Improved Transformer with Multi-scale Attention | 91.6% accuracy | 6.1% increase |
| Chemical Engineering Construction [6] | Conventional Machine Learning | ~76.3% accuracy | Improved Transformer with Data Fusion | 91.6% accuracy | 19.4% increase |
The experimental data consistently demonstrates that appropriately implemented data fusion strategies outperform single-technique approaches across multiple domains. For herbal extract monitoring, high-level data fusion integrating FT-NIR and Vis/NIR-HSI achieved superior prediction accuracy for all critical quality attributes compared to either technique alone [57]. Similarly, in complex chemical engineering environments, improved Transformer architectures with multi-scale attention mechanisms significantly outperformed both conventional machine learning and standard Transformer models [6].
The Scientist's Toolkit: Essential Research Reagents and Solutions
Table 3 catalogues key reagents, computational tools, and methodologies referenced in the experimental studies:
Table 3: Research Reagent Solutions for Data Fusion Experiments
| Item | Function/Application | Specification/Implementation |
|---|---|---|
| Fourier Transform Near-Infrared (FT-NIR) Spectroscopy | Non-destructive quantification of chemical compounds and moisture content [57] | Spectral range: 800-2500 nm; Used with partial least squares regression |
| Visible/Near-Infrared Hyperspectral Imaging (Vis/NIR-HSI) | Simultaneous capture of spatial and spectral information for physical and chemical properties [57] | Spectral range: 400-1000 nm; Provides both spatial and quantitative data |
| Adaptive Weight Allocation Algorithm | Dynamically adjusts data source contributions based on real-time quality assessment [6] | Improved Transformer architecture; Addresses varying data reliability in industrial environments |
| Penalized Cox Partial Likelihood | Identifies homogeneous and heterogeneous effects in pooled survival data [50] | Adaptive group lasso with composite penalties; Implemented via iterative shooting algorithm |
| Multi-scale Attention Mechanism | Models temporal hierarchies in processes with different sampling frequencies [6] | Domain-specific Transformer extension; Handles millisecond sensor readings to monthly reports |
| Partial Least Squares (PLS) Regression | Builds predictive models when predictors are highly collinear or numerous [57] | Used for spectral data modeling; Implementation with data fusion strategies |
Integrated Workflow for Mitigating Pitfalls in Pharmaceutical Research
Successfully addressing data heterogeneity, dimensionality, and overfitting requires a systematic approach that incorporates advanced computational techniques while maintaining biological relevance. Figure 2 illustrates an integrated workflow for pharmaceutical research applications:
Figure 2: Integrated Workflow for Managing Data Challenges in Pharmaceutical Research
The experimental evidence demonstrates that data fusion strategies systematically outperform single-technique approaches in predictive accuracy across multiple domains, with documented improvements of 6.1–19.4% in various applications [57] [6]. However, these advantages come with implementation complexities, including increased computational demands and the need for specialized expertise.
For drug development professionals, the choice between approaches involves careful consideration of specific research contexts. Single-technique methods may suffice for well-defined problems with homogeneous data sources, while data fusion approaches become indispensable for integrating diverse data modalities and managing inherent heterogeneity [57] [50] [6]. Critically, successful implementation requires appropriate dimensionality reduction to mitigate overfitting risks in high-dimensional spaces [51] [56].
Future advancements in Transformer architectures with domain-specific attention mechanisms [6] and adaptive regularization methods for heterogeneous data [50] promise to further enhance data fusion capabilities. By strategically selecting methodologies that address all three pitfalls—heterogeneity, dimensionality, and overfitting—researchers can develop more robust, generalizable predictive models to accelerate pharmaceutical innovation.
In the pursuit of enhanced predictive accuracy in data-driven research, the integration of multimodal data has emerged as a pivotal strategy. This guide objectively compares two principal data fusion strategies—Early Fusion and Late Fusion—within the broader context of academic thesis research aimed at surpassing the performance limitations of single-technique models. The selection between these paradigms is not merely an implementation detail but a critical methodological determinant of model accuracy, robustness, and generalizability. Recent theoretical and applied research provides a framework for making this choice a priori, based on dataset characteristics and project constraints, thereby optimizing computational resources and outcome reliability [2].
Core Concepts and Definitions
Early Fusion
Early Fusion (EF), or feature-level fusion, involves the integration of raw or pre-processed features from multiple data sources into a single, combined feature vector before the data is input into a machine learning model.
- Mechanism: Features from
Kmodalities are concatenated or pooled into one unified dataset. A single model, often a generalized linear model, is then trained on this fused dataset to learn patterns that incorporate inter-modal relationships [2] [58]. - Theoretical Foundation: Under the framework of generalized linear models, EF can be formally defined as a model where a connection function
g_Esatisfiesg_E(μ) = η_E = Σ w_i * x_i, wherex_iare features from all modalities andw_iare their corresponding non-zero weight coefficients [2].
Late Fusion
Late Fusion (LF), also known as decision-level fusion, employs separate models for each data modality. The final prediction is achieved by aggregating the decisions or prediction probabilities from these individual models.
- Mechanism: Individual models are trained independently on their respective modalities. Their outputs are then combined using an aggregation function
f, which can be averaging, weighted voting, or a meta-classifier [2] [58]. - Theoretical Foundation: For
Kmodalities, LF is defined by sub-modelsg_Lk(μ) = η_Lkfor each modalityk, with the final output beingoutput_L = f( g_L1^{-1}(η_L1), ..., g_LK^{-1}(η_LK) )[2].
Comparative Performance Analysis Across Domains
The following table synthesizes quantitative findings from recent peer-reviewed studies, comparing the performance of Early and Late Fusion strategies across diverse scientific fields.
Table 1: Comparative Performance of Early vs. Late Fusion in Recent Research
| Research Domain | Specific Task | Early Fusion Performance | Late Fusion Performance | Key Performance Metric | Source |
|---|---|---|---|---|---|
| Dementia Care | Aggression detection from audio & visual signals | Accuracy: 0.828Precision: 0.852Recall: 0.818F1-Score: 0.835ROC-AUC: 0.922 | Accuracy: 0.876Precision: 0.824Recall: 0.914F1-Score: 0.867ROC-AUC: 0.970 | Multiple | [59] |
| Oncology | Breast cancer survival prediction | Lower test-set concordance | Consistently higher test-set concordance | Concordance Index | [34] |
| Neuroimaging | Alzheimer's disease classification (sMRI & MEG) | Accuracy: 0.756 | Accuracy: 0.801 | Accuracy | [60] |
| Plant Breeding | Genomic & Phenotypic Selection (GPS) | Highest accuracy (53.4% improvement over best single model) | Lower accuracy than data fusion | Predictive Accuracy | [3] |
| Cardiology | Heart disease prediction (CNN & DNN) | Lower accuracy than late fusion | Accuracy, Precision, Recall, F1: ~99.99% | Multiple | [61] |
Experimental Protocols and Methodologies
To ensure the reproducibility of findings cited in this guide, this section details the experimental protocols from key studies.
- Objective: To classify aggression using audio and visual signals from dementia patients.
- Data Modalities: Audio recordings and visual data.
- Fusion Implementation:
- Early Fusion: Extracted features from both modalities were concatenated into a single dataset prior to classification.
- Late Fusion: Standalone classifiers were trained on audio and visual data separately. The prediction probabilities from both models were then integrated using a meta-classifier.
- Model & Validation: A Random Forest classifier was used for both fusion strategies. Model performance was evaluated using 5-fold cross-validation, and a paired t-test was applied for statistical significance.
- Objective: To predict overall survival by integrating clinical, multi-omics, and histopathology imaging data.
- Data Modalities: Clinical variables, somatic mutations, RNA expression, copy number variation, miRNA expression, and whole-slide histopathology images from TCGA.
- Fusion Implementation:
- Early Fusion: Features from all modalities were simply concatenated at the input layer.
- Late Fusion: Separate models were trained for each modality. Their predictions were combined at the decision level.
- Model & Validation: A rigorous validation framework was employed, using cross-validation and a fixed, held-out test set to ensure an unbiased assessment of out-of-sample generalization performance.
- Objective: To enhance predictive accuracy for complex traits in plant breeding.
- Data Modalities: Genomic data and phenotypic data.
- Fusion Strategies Tested:
- Data Fusion (Early): Integration of raw genomic and phenotypic data before modeling.
- Feature Fusion (Intermediate): Fusion of learned features from each data type.
- Result Fusion (Late): Aggregation of predictions from separate genomic and phenotypic models.
- Model & Validation: The framework was tested on large datasets from four crop species using a suite of models, demonstrating robustness across sample sizes and environments.
Visualizing Fusion Architectures
The fundamental workflows for Early and Late Fusion are illustrated below, highlighting the key differences in data handling and model architecture.
Diagram 1: A comparison of Early and Late Fusion data workflows, showing the integration point of multimodal data.
The Selection Paradigm: A Decision Framework
The choice between Early and Late Fusion is not arbitrary. Research indicates that the optimal strategy depends on specific dataset characteristics and project goals. The following diagram outlines a evidence-based decision paradigm.
Diagram 2: A practical decision framework for selecting between Early and Late Fusion, based on project constraints.
This paradigm is supported by key research findings:
- Sample Size is a Critical Factor: Theoretical work has proposed a critical sample size threshold at which the performance dominance of early and late fusion reverses. With small sample sizes, Late Fusion consistently demonstrates superior generalization, as seen in breast cancer survival prediction [2] [34]. Early Fusion requires large amounts of data to avoid overfitting in its high-dimensional feature space [2] [26].
- Inter-Modality Relationships: Early Fusion excels when strong, complementary relationships exist between features of different modalities, as it allows the model to learn these interactions directly [2] [26]. Conversely, Late Fusion is more robust when such relationships are weak or nonlinear [2].
- System Flexibility and Generalization: If the research project requires modularity (e.g., plans to incorporate new data types later) or prioritizes model generalization on unseen data, Late Fusion is the preferred choice [59] [34].
The Scientist's Toolkit: Essential Research Reagents and Solutions
The following table details key computational tools and resources essential for implementing and testing data fusion strategies in a research environment.
Table 2: Key Research Reagents and Solutions for Data Fusion Experiments
| Tool/Resource | Type/Category | Primary Function in Fusion Research | Exemplary Use Case |
|---|---|---|---|
| Random Forest Classifier | Machine Learning Algorithm | A versatile model for classification tasks; used as a base classifier in both EF and LF, and as a meta-classifier in LF aggregation. | Used as the primary classifier and meta-classifier in multimodal aggression detection [59]. |
| XGBoost | Machine Learning Algorithm | A powerful gradient-boosting algorithm effective for structured/tabular data; can serve as a predictive model in EF or as an expert model per modality in LF. | Top-performing model for global sea surface nitrate estimation, outperforming six other algorithms [4]. |
| Convolutional Neural Network (CNN) | Deep Learning Architecture | Extracts spatial features from image or signal data (e.g., spectrograms); acts as a modality-specific expert model in LF frameworks. | Used with DNNs in a late fusion framework for heart disease prediction, achieving near-perfect metrics [61]. |
| The Cancer Genome Atlas (TCGA) | Biomedical Dataset | Provides large-scale, multi-modal data (clinical, genomic, image) essential for training and validating fusion models in oncology. | Served as the data source for clinical, omics, and histopathology images in breast cancer survival prediction [34]. |
| Cross-Validation Framework | Statistical Methodology | Ensures robust performance estimation and mitigates overfitting; critical for fairly comparing EF and LF generalization error. | Employed via 5-fold cross-validation in dementia aggression studies and breast cancer survival prediction [59] [34]. |
In the evolving landscape of data-driven research, a critical phenomenon has emerged: the reversal of performance dominance between predictive methodologies at specific sample size thresholds. This paradigm challenges conventional wisdom in experimental design, particularly for researchers, scientists, and drug development professionals who rely on accurate predictive modeling. While data fusion techniques integrate multiple data sources to enhance predictive accuracy, and single-technique approaches offer simplicity and interpretability, their relative performance is not static but fluctuates dramatically with sample size variations [2]. Understanding these performance reversal thresholds is essential for optimizing research outcomes and resource allocation.
The fundamental relationship between sample size and predictive accuracy represents a pivotal consideration across scientific disciplines. Evidence indicates that robust reliability estimation often requires larger samples than conventionally used, with some behavioral and computational measures needing 150-300 participants for stable variance component estimation [62]. Simultaneously, adaptive designs with sample size recalculation have demonstrated improved power maintenance in randomized test-treatment trials [63]. This article examines the critical intersection where data fusion and single-technique research methodologies undergo performance reversals, providing evidence-based guidance for experimental design decisions in pharmaceutical and scientific research contexts.
Theoretical Foundations: Data Fusion Methods and Performance Metrics
Data Fusion Classification Frameworks
Data fusion methodologies can be systematically categorized based on processing level and architectural approach. The Dasarathy classification system provides a foundational framework, distinguishing five categories based on input/output data types: Data In-Data Out (DAI-DAO), Data In-Feature Out (DAI-FEO), Feature In-Feature Out (FEI-FEO), Feature In-Decision Out (FEI-DEO), and Decision In-Decision Out (DEI-DEO) [24]. These categories represent increasing levels of abstraction, from raw data combination to symbolic decision integration.
Alternative classification approaches include:
- Durrant-Whyte's framework categorizes fusion based on source relationships: complementary (different parts of scene), redundant (same target for confidence increase), and cooperative (generating new complex information) [24]
- Abstraction-level classification distinguishes signal, pixel, characteristic, and symbol levels [24]
- JDL model conceptualizes five processing levels: source preprocessing, object refinement, situation assessment, impact assessment, and process refinement [24]
Contemporary Fusion Paradigms
Three primary fusion architectures dominate current research applications:
Early Fusion (Data Layer): Concatenates raw features from multiple modalities as input to a single predictive model. This approach preserves potential cross-modal interactions but increases dimensionality [2].
Late Fusion (Decision Layer): Trains separate models on different data modalities and aggregates their predictions. This method accommodates modality-specific processing but may miss cross-modal correlations [2].
Gradual/Intermediate Fusion: Processes modalities stepwise according to correlation strength, offering a balanced approach between early and late fusion [2].
The Sample Size Performance Reversal Phenomenon
Mathematical Foundations of Performance Reversal
The relationship between sample size and predictive accuracy follows quantifiable patterns that explain performance reversal phenomena. Under the generalized linear model framework, the performance dominance between early fusion and late fusion reverses at a critical sample size threshold [2]. This reversal occurs because early fusion models, with their higher parameter counts, require sufficient data to overcome the curse of dimensionality, while late fusion methods demonstrate greater efficiency with limited data but eventually plateau in performance.
Mathematical analysis reveals that early fusion connection functions take the form:
where all features are concatenated into a single vector [2]. In contrast, late fusion employs separate sub-models for each modality:
with decision aggregation:
The different learning trajectories of these architectures create the reversal phenomenon [2].
Empirical Evidence from Multidisciplinary Studies
Table 1: Sample Size Requirements for Reliability Estimation Across Disciplines
| Field/Application | Reliability Metric | Required Sample Size | Key Findings |
|---|---|---|---|
| Behavioural Reliability Research | Intraclass Correlation Coefficient (ICC) | 10 to >300 participants | Between-subject variance required median N=167; within-subject variance required median N=34 [62] |
| Computational Psychiatry | Test-retest reliability of reversal learning parameters | Median N=68 between-subject, N=20 within-subject, N=45 error variance [62] | Sample sizes exceeding typical reliability studies (circa N=30) needed for robust estimates [62] |
| Plant Breeding (GPS Framework) | Genomic and Phenotypic Selection accuracy | Sample size as small as N=200 maintained high predictive accuracy [3] | Lasso_D data fusion achieved 53.4% accuracy improvement over best genomic selection model [3] |
| Randomized Test-Treatment Trials | Adaptive design with blinded sample size recalculation | Varies based on interim overall success rate re-estimation [63] | Maintained theoretical power with unbiased estimates compared to over/under-powered fixed designs [63] |
Evidence from reliability research demonstrates that variance component estimation requires different sample sizes for different sources of variance, with between-subject variance generally demanding the largest samples [62]. This has direct implications for fusion methodologies, as late fusion may better handle high between-subject variance with limited samples.
In plant breeding applications, the GPS framework (genomic and phenotypic selection) demonstrated that data fusion maintained robust predictive accuracy even with relatively small sample sizes (N=200), outperforming single-technique approaches by significant margins (18.7-53.4% accuracy improvements) [3]. This highlights the potential for data fusion to mitigate sample size constraints in specific applications.
Experimental Protocols and Methodologies
Reliability Estimation and Variance Decomposition
Quantifying test-retest reliability requires careful experimental design and statistical analysis. The intraclass correlation coefficient (ICC) serves as a standard metric for assessing a measure's ability to quantify systematic between-subject differences [62]. However, ICC calculations are more heavily influenced by increases in between-subject variance than session variance effects, potentially introducing bias [62].
Variance decomposition addresses this limitation by maintaining variance components in composite parts: within-subject session variance, between-subject variance, and error variance [62]. The experimental protocol involves:
- Repeated measurements across multiple sessions with identical protocols
- Large sample recruitment to ensure stable variance component estimation
- Simulation approaches based on ground truth data to investigate sample size effects
- Stability assessment using precision (half-width) and confidence (point of stability percentile) metrics
For reversal learning tasks as assays of behavioral flexibility, studies have established reliability through test-retest designs with computational modeling to derive latent descriptors of performance [62].
Data Fusion Experimental Framework
The comparative evaluation of data fusion methods follows standardized protocols:
Modality Segmentation: Features are organized into modality-specific sets ( Xi = (x1^i, x2^i, ..., x{m_i}^i) ) for ( i = 1 ) to ( K ) modalities [2]
Model Specification: Generalized linear models are implemented with appropriate link functions connecting linear predictors to response variable expectations [2]
Fusion Implementation:
- Early fusion: All features concatenated into single input vector
- Late fusion: Separate models per modality with decision aggregation
- Gradual fusion: Stepwise fusion based on inter-modal correlations
Performance Assessment: Threshold-free evaluation using Area Under the Curve metrics to avoid selection bias [64]
Table 2: Key Research Reagent Solutions for Data Fusion Experiments
| Research Reagent | Function/Purpose | Application Context |
|---|---|---|
| Generalized Linear Models (GLM) | Provides mathematical framework relating independent variables to expected response values through link functions [2] | Fundamental modeling approach for all fusion methodologies |
| Pooled Adjacent Violators Algorithm (PAVA) | Fits isotonic regression for monotonic dose-response relationships without parametric assumptions [65] | Threshold value estimation in medical and toxicological studies |
| Lasso_D (Data Fusion) | Regression method with L1 regularization for high-dimensional data in fusion contexts [3] | Plant breeding genomic and phenotypic selection |
| Intraclass Correlation Coefficient (ICC) | Quantifies test-retest reliability by partitioning variance components [62] | Behavioral and computational reliability assessment |
| AUC-IoU (Area Under Curve - Intersection over Union) | Threshold-free evaluation metric for attribution methods eliminating selection bias [64] | Comparative assessment of fusion method performance |
Adaptive Sample Size Recalculation Protocol
Randomized test-treatment studies with restricted randomization to discordant pairs employ specific adaptive designs:
- Pilot Phase: Initial data collection with blinded conditions
- Interim Analysis: Re-estimation of nuisance parameters (e.g., overall success rate for binary endpoints)
- Sample Size Recalculation: Adjustment of total sample size based on re-estimated parameters
- Final Analysis: Incorporation of all observations from both phases [63]
This approach maintains type I error control while improving power compared to fixed designs, addressing the critical challenge of uncertain assumptions in trial planning [63].
Comparative Performance Analysis
Sample Size Thresholds for Method Selection
The performance dominance reversal between early and late fusion occurs at a mathematically definable critical sample size threshold [2]. This threshold depends on three key parameters:
- Feature dimensionality across modalities
- Correlation strength between auxiliary traits and target traits
- Model complexity and regularization approaches
Experimental results demonstrate that early fusion typically outperforms late fusion with sufficient sample sizes, but its performance degrades more rapidly with sample size reduction due to increased parameterization [2]. The GPS framework in plant breeding achieved particularly impressive results, with Lasso_D data fusion improving selection accuracy by 53.4% compared to the best genomic selection model and by 18.7% compared to the best phenotypic selection model [3].
Reliability and Stability Considerations
The relationship between sample size and reliability estimation quality follows distinct patterns for different variance components. Research indicates that with ±0.05 precision and 80% confidence, stable estimates require:
- Between-subject variance: Largest samples (median N=167 for behavioral measures, N=68 for computational measures)
- Within-subject variance: Moderate samples (median N=34 for behavioral, N=20 for computational)
- Error variance: Intermediate samples (median N=103 for behavioral, N=45 for computational) [62]
These differential requirements have direct implications for fusion methodology selection in reliability studies, particularly when specific variance components dominate the measurement construct.
Visualization of Methodologies and Relationships
Data Fusion Workflow Comparison
Data Fusion Method Workflows
Sample Size Performance Reversal Visualization
Performance Reversal Across Sample Size Regimes
The critical sample size phenomenon represents a fundamental consideration in the comparative evaluation of data fusion versus single-technique predictive approaches. Performance reversal thresholds dictate methodological selection, with late fusion generally preferable in limited sample regimes and early fusion increasingly advantageous as sample sizes surpass critical thresholds. For research and drug development professionals, these insights enable evidence-based experimental design decisions, optimizing predictive accuracy while efficiently allocating resources. Future research should continue to refine mathematical models of these reversal thresholds across diverse application domains, particularly as data fusion methodologies evolve in complexity and applicability.
Optimizing Computational Efficiency and Model Robustness
In the data-intensive fields of modern drug development and chemical engineering, the choice between using a single data source and integrating multiple ones through data fusion is a critical determinant of a model's predictive accuracy, computational efficiency, and ultimate robustness. Data fusion, the multi-level process of associating, correlating, and combining data from single and multiple sources, has emerged as a pivotal methodology for achieving refined estimations and complete assessments [2]. The central thesis of this guide is that while single-source models offer simplicity, advanced data fusion methods—namely early, late, and gradual fusion—systematically enhance predictive performance for complex biological and chemical problems, albeit with distinct computational trade-offs that must be optimized for specific research contexts.
The pharmaceutical and biotechnology sectors, where AI spending is projected to reach $3 billion by 2025, present a compelling use-case for these methodologies [66]. In drug discovery, the traditional single-technique approach often struggles with the multifactorial nature of disease mechanisms and drug-target interactions. Consequently, multimodal data fusion enables researchers to integrate diverse data streams—from genomic profiles and molecular structures to clinical trial outcomes and real-world evidence—creating predictive models that more accurately simulate complex biological reality [67] [68]. This guide provides a structured comparison of dominant fusion methodologies, supported by experimental data and practical implementation protocols, to equip researchers with evidence-based selection criteria for their specific computational challenges.
Theoretical Foundations of Data Fusion Methods
Data fusion strategies are broadly classified into three architectural paradigms based on the stage at which integration occurs: early fusion, late fusion, and gradual fusion. Each employs distinct mechanisms for combining information and consequently exhibits characteristic performance profiles.
Early Fusion (Data-Level Fusion) operates at the most basic level by simply concatenating raw features from multiple modalities into a single input vector for a predictive model. Within the framework of generalized linear models, early fusion can be mathematically represented as gE(μ) = ηE = Σ(i=1 to m) wixi, where gE is the connection function, ηE is the linear predictor output, wi are the non-zero weight coefficients, and xi are the features from all modalities [2]. The final prediction is obtained by applying the inverse of the link function, gE^{-1}(ηE). This approach preserves potential inter-modal dependencies at the cost of increased dimensionality.
Late Fusion (Decision-Level Fusion) employs a distributed approach where separate models are trained independently on each data modality. The predictions from these unimodal models are then aggregated through a fusion function. Formally, for K modes, each sub-model follows gLk(μ) = ηLk = Σ(j=1 to mk) wjk xjk, where xjk ∈ Xk [2]. The final decision is computed as outputL = f(gL1^{-1}(ηL1), gL2^{-1}(ηL2), ..., gLK^{-1}(ηLK)), where f is the fusion function, which could be a weighted average, voting mechanism, or another meta-learner.
Gradual Fusion (Intermediate/Feature-Level Fusion) processes data through a hierarchical, stepwise manner according to the correlation between different modalities, with highly correlated modes fused earlier than those with lower correlation [2]. This approach can be represented as gG(μ) = ηG = G(X̄, F), where X̄ represents the set of all modal features and F represents the set of fusion prediction functions arranged in a network graph that defines the input relations and fusion sequence.
Table 1: Theoretical Comparison of Data Fusion Architectures
| Fusion Method | Integration Level | Mathematical Formulation | Key Mechanism |
|---|---|---|---|
| Early Fusion | Data/Input Level | gE(μ) = ηE = Σ(i=1 to m) wixi |
Concatenation of raw features into unified input vector |
| Late Fusion | Decision/Output Level | outputL = f(gL1^{-1}(ηL1), ..., gLK^{-1}(ηLK)) |
Aggregation of decisions from specialized unimodal models |
| Gradual Fusion | Feature/Intermediate Level | gG(μ) = ηG = G(X̄, F) |
Stepwise hierarchical fusion based on inter-modal correlations |
Experimental Comparison: Performance Across Domains
Quantitative Performance Benchmarks
Rigorous experimental evaluations across multiple domains demonstrate the consistent superiority of data fusion approaches over single-source methods, with the optimal fusion strategy being highly context-dependent.
In spectroscopic analysis of industrial lubricant additives and minerals, a novel Complex-level Ensemble Fusion (CLF) method significantly outperformed both single-source models and classical fusion schemes. This two-layer chemometric algorithm jointly selects variables from concatenated mid-infrared (MIR) and Raman spectra using a genetic algorithm, projects them with partial least squares, and stacks the latent variables into an XGBoost regressor [69]. The CLF approach effectively captured feature- and model-level complementarities in a single workflow, achieving predictive accuracy unattainable by any single-spectral technique.
For chemical engineering construction projects, an improved Transformer architecture with enhanced attention mechanisms achieved remarkable performance in multi-source heterogeneous data fusion. The system integrated structured numerical measurements, semi-structured operational logs, and unstructured textual documentation through multi-scale attention mechanisms and cross-modal feature alignment modules [6]. Experimental validation demonstrated prediction accuracies exceeding 91% across multiple tasks, representing improvements of up to 19.4% over conventional machine learning techniques and 6.1% over standard Transformer architectures.
Table 2: Experimental Performance Comparison of Data Fusion vs. Single-Source Methods
| Application Domain | Single-Source Baseline Accuracy | Optimal Fusion Method | Fusion Accuracy | Performance Gain |
|---|---|---|---|---|
| Spectroscopic Analysis (Industrial Lubricants) | 72-78% (MIR or Raman only) | Complex-level Ensemble Fusion (CLF) | Significantly outperformed all baselines | >15% improvement over best single-source |
| Chemical Engineering Construction | ~76% (Traditional ML) | Improved Transformer with Attention | >91% | 19.4% improvement |
| Wind Turbine Blade Assessment | Limited to local comparisons | Full-Field Data Fusion (FFDF) | Enabled quantitative full-field validation | Previously unattainable metrics |
| Drug Discovery Timeline | 10-15 years (Traditional) | AI-driven Multi-modal Fusion | 12-18 months for candidate identification | ~50% reduction [70] |
Cross-Domain Methodological Efficacy
Beyond specific accuracy metrics, data fusion demonstrates transformative potential across research domains by enabling analyses previously impossible with single-source approaches.
In wind turbine blade substructure evaluation, Full-Field Data Fusion (FFDF) methodology combined Digital Image Correlation (DIC) and Thermoelastic Stress Analysis (TSA) experimental data with Finite Element Analysis (FEA) predictions [71]. This approach enabled quantitative full-field comparisons and created novel performance parameters unattainable by any single technique, such as combined strain-stress-stiffness metrics. The fusion eliminated inaccuracies from comparable location estimation and enabled mutual assessment of experimental technique reliability.
For pharmaceutical R&D, AI-driven fusion of multimodal data has dramatically compressed development timelines. Companies like Insilico Medicine have demonstrated the ability to identify novel drug candidates for complex diseases like idiopathic pulmonary fibrosis in approximately 18 months—a fraction of the traditional 3-6 year timeline for this stage [68]. This acceleration stems from fusion-driven integration of chemical, biological, and clinical data to simultaneously optimize multiple drug properties.
Selection Framework and Implementation Protocols
Fusion Method Selection Paradigm
Research indicates that the optimal fusion strategy depends on specific dataset characteristics and problem constraints. A comparative analysis of early, late, and gradual fusion methods has yielded a structured selection paradigm based on sample size, feature quantity, and modality relationships [2].
Critical findings from this theoretical analysis include equivalence conditions between early and late fusion within generalized linear models, failure conditions for early fusion in the presence of nonlinear feature-label relationships, and an approximate equation for evaluating the accuracy of early versus late fusion as a function of sample size, feature quantity, and modality number. Most significantly, researchers identified a critical sample size threshold at which performance dominance reverses between early and late fusion approaches [2].
For problems with limited samples and high-dimensional features, late fusion typically outperforms early fusion by avoiding the curse of dimensionality. As sample size increases beyond the critical threshold, early fusion potentially leverages inter-modal dependencies more effectively. Gradual fusion emerges as the superior approach when modalities exhibit varying correlation patterns and when computational resources allow for hierarchical processing.
Implementation Protocols for Featured Studies
Protocol 1: Complex-level Ensemble Fusion for Spectroscopic Data
The CLF methodology implemented a two-layer architecture for fusing mid-infrared and Raman spectroscopic data [69]:
Variable Selection: A genetic algorithm jointly selected informative variables from concatenated MIR and Raman spectra, identifying complementary spectral regions.
Feature Projection: Selected variables underwent projection via Partial Least Squares to latent structures, reducing dimensionality while preserving predictive information.
Ensemble Stacking: Latent variables from both modalities were stacked as input to an XGBoost regressor, which learned optimal integration weights through boosted tree ensemble.
Validation: The approach employed rigorous cross-validation against single-source models and classical low-, mid-, and high-level fusion schemes on industrial lubricant additives and RRUFF mineral datasets.
Protocol 2: Transformer-based Multi-source Fusion for Chemical Engineering
The improved Transformer architecture for chemical engineering construction projects implemented these key methodological innovations [6]:
Multi-scale Attention Mechanism: Domain-specific attention explicitly modeled temporal hierarchies in construction processes, handling data streams with vastly different sampling frequencies.
Cross-modal Alignment: A contrastive framework learned semantic correspondences between numerical sensor data, textual documentation, and categorical project states without manual feature engineering.
Adaptive Weight Allocation: A dynamic algorithm adjusted data source contributions based on real-time quality assessment and task-specific relevance.
Multi-task Learning: The architecture simultaneously predicted progress estimation, quality assessment, and risk evaluation while maintaining task-specific adaptation capabilities.
The Scientist's Toolkit: Essential Research Reagents & Computational Solutions
Table 3: Essential Research Reagents and Computational Solutions for Data Fusion Experiments
| Tool/Reagent | Function/Role | Application Context |
|---|---|---|
| Transformer Architectures | Multi-scale attention for temporal hierarchy modeling; cross-modal alignment | Chemical engineering construction projects; multi-source heterogeneous data [6] |
| Complex-level Ensemble Fusion | Two-layer chemometric algorithm combining genetic algorithm variable selection with ensemble stacking | Spectroscopic data fusion (MIR & Raman); industrial quality control [69] |
| Full-Field Data Fusion Framework | Quantitative comparison of full-field experimental techniques via spatial resolution unification | Wind turbine blade assessment; experimental mechanics validation [71] |
| AlphaFold & Generative AI | Protein structure prediction; molecular design and optimization | Drug discovery; target identification and compound screening [67] [68] |
| XGBoost Regressor | Ensemble learning for final prediction layer in stacked fusion architectures | Spectroscopic data fusion; chemical property prediction [69] |
| Digital Image Correlation | Surface displacement and strain measurement via optical pattern tracking | Experimental mechanics; structural validation [71] |
| Thermoelastic Stress Analysis | Surface stress measurement via infrared detection of thermoelastic effect | Experimental mechanics; stress distribution analysis [71] |
The evidence from multiple research domains consistently demonstrates that strategic data fusion implementation enhances both predictive accuracy and model robustness compared to single-technique approaches. The key to optimizing computational efficiency lies in method selection matched to specific research constraints—particularly sample size, feature dimensionality, and inter-modal relationships.
For drug development professionals, these findings translate to tangible efficiencies: AI-driven data fusion has demonstrated potential to reduce drug discovery timelines from years to months while improving success rates [70] [68]. In chemical engineering and materials science, fusion methodologies enable more comprehensive system evaluations through integrated analysis of previously siloed data streams [6] [71].
The future trajectory points toward increasingly sophisticated fusion architectures, particularly improved Transformer models with domain-specific attention mechanisms and automated fusion strategy selection. As these methodologies mature, researchers who strategically implement data fusion protocols will gain significant advantages in predictive capability, resource efficiency, and ultimately, research impact across pharmaceutical, chemical, and engineering domains.
Proof of Concept: Validating and Comparing Fusion Against Single Techniques
Establishing Rigely, Rigorous Evaluation Metrics for Multimodal Models
In the high-stakes domain of drug development, the transition from unimodal to multimodal artificial intelligence (AI) represents a paradigm shift comparable to the move from single-target therapies to systems biology. While traditional AI models excel within their specialized domains—processing text, images, or molecular structures in isolation—they fundamentally lack the integrative capacity to model complex biological systems. Multimodal AI, which processes and correlates diverse data types within a unified model, offers a transformative approach by mirroring the multi-faceted nature of biological reality itself. The core thesis underpinning this evolution posits that strategic data fusion significantly enhances predictive accuracy beyond the capabilities of any single-technique approach, potentially accelerating therapeutic discovery and reducing costly late-stage failures.
This guide provides a rigorous framework for evaluating multimodal models, with particular emphasis on applications in pharmaceutical research and development. We objectively compare leading platforms through the lens of domain-specific performance metrics, experimental protocols, and practical implementation considerations, providing scientists and researchers with the analytical tools necessary to validate and deploy these advanced systems.
Theoretical Foundations: Data Fusion Strategies and Their Implications
Fusion Methodologies: A Comparative Framework
The architectural approach to combining disparate data types fundamentally influences model performance, robustness, and interpretability. Research analyzing data fusion methods within generalized linear models has established mathematical conditions under which different fusion strategies excel [2]. The three primary fusion methodologies are:
Early Fusion (Data-Level Fusion): This approach involves concatenating raw or pre-processed features from multiple modalities into a single input vector before model training [2]. For example, combining molecular structures, transcriptomic profiles, and clinical trial outcomes into a unified feature set. Theoretically, early fusion can capture complex, nonlinear interactions between modalities at the cost of increased dimensionality and potential overfitting on limited biomedical datasets.
Late Fusion (Decision-Level Fusion): In this architecture, separate unimodal models are trained independently on their respective data types, with their predictions combined at the decision layer through ensemble methods or learned weighting functions [2]. This approach preserves modality-specific feature relationships while requiring careful design of the fusion function to optimize complementary information.
Intermediate (Gradual) Fusion: This hybrid approach processes modalities through separate encoders before fusing representations at intermediate network layers [2]. This enables the model to learn both modality-specific and cross-modal representations, offering a balance between the interaction modeling of early fusion and the robustness of late fusion.
Theoretical Performance Dominance Conditions
Mathematical analysis reveals that the performance dominance between early and late fusion is not absolute but contingent on dataset characteristics and learning parameters. Research has established that a critical sample size threshold exists at which the performance superiority reverses between early and late fusion approaches [2]. Specifically:
Early fusion dominance conditions: Early fusion generally outperforms when dealing with large sample sizes relative to feature dimensions and when strong nonlinear interactions exist between modalities. This makes it particularly suitable for problems with abundant multimodal data where capturing complex cross-modal interactions is essential.
Late fusion dominance conditions: Late fusion demonstrates superior performance with limited training samples or when modalities exhibit weak interdependence. This advantage stems from its reduction of the hypothesis space by training separate models on each modality, thereby mitigating the curse of dimensionality.
The mathematical derivation of this critical threshold provides researchers with a principled basis for selecting fusion strategies prior to model development, potentially saving substantial computational resources [2].
Evaluation Benchmarks and Performance Metrics
Comprehensive Benchmarking Frameworks
Standardized benchmarks enable objective comparison of multimodal models across diverse capabilities. The most comprehensive evaluation frameworks assess both perceptual abilities (identifying elements in data) and cognitive capabilities (reasoning about identified elements) [72].
Table 1: Foundational Multimodal Model Evaluation Benchmarks
| Benchmark Name | Primary Focus | Modalities | Key Metrics | Domain Relevance |
|---|---|---|---|---|
| MME | Comprehensive perception & cognition evaluation | Vision, Language | Accuracy across 14 subtasks | General capability assessment [72] |
| TrialBench | Clinical trial outcome prediction | Molecular, Clinical, Text | AUC-ROC, Accuracy, F1-Score | Drug development specific [73] |
| GAIA | General AI assistant capabilities | Mixed real-world tasks | Task success rate | Practical application testing [74] |
| AgentBench | Agent-based task performance | Web, OS, Database | Success rate, Efficiency | Tool-use and decision making [74] |
For drug development applications, specialized benchmarks like TrialBench provide clinically-relevant evaluation frameworks. This comprehensive suite comprises 23 AI-ready datasets addressing 8 critical clinical trial prediction challenges: trial duration, patient dropout, adverse events, mortality, trial approval, failure reasons, drug dosage, and eligibility criteria design [73].
Domain-Specific vs. Generic Evaluation Metrics
In biomedical applications, standard ML metrics often prove inadequate for capturing model performance in real-world contexts. The limitations of conventional metrics become particularly apparent when dealing with imbalanced datasets common in drug discovery, where inactive compounds vastly outnumber active ones [75].
Table 2: Metric Comparison for Drug Discovery Applications
| Metric Category | Standard Metric | Domain-Specific Adaptation | Application Context |
|---|---|---|---|
| Accuracy Measures | F1-Score | Precision-at-K | Prioritizing top drug candidates [75] |
| Event Detection | Accuracy | Rare Event Sensitivity | Identifying adverse drug reactions [75] |
| Biological Relevance | ROC-AUC | Pathway Impact Metrics | Assessing biological mechanism alignment [75] |
| Model Robustness | Cross-Validation Score | Multi-Modal Consistency Check | Evaluating cross-domain performance [76] |
Domain-specific metrics address the critical need for biologically meaningful evaluation in pharmaceutical applications. For example, Pathway Impact Metrics assess how well model predictions align with known biological pathways, ensuring computational findings translate to mechanistically plausible insights [75]. Similarly, Rare Event Sensitivity specifically measures a model's ability to detect low-frequency but high-consequence events like serious adverse drug reactions, which conventional accuracy metrics might overlook despite their clinical significance [75].
Comparative Analysis of Leading Multimodal Platforms
Performance Across Technical Specifications
Objective comparison of multimodal platforms requires examination across multiple dimensions, including architectural capabilities, performance benchmarks, and practical implementation considerations.
Table 3: Multimodal Platform Comparison for Scientific Applications
| Platform | Context Capacity | Modalities Supported | Key Strengths | Domain-Specific Performance |
|---|---|---|---|---|
| GPT-4o | 128K tokens | Text, Image, Audio | Real-time multimodal interaction, Native audio understanding | Limited by 4K output for document-heavy workflows [77] |
| Gemini 2.5 Pro | 2M tokens | Text, Image, Video, Audio | Massive context for codebases/long documents | 92% accuracy on benchmarks; Legal document review [77] |
| Claude Opus | 200K tokens | Text, Image | Constitutional training for safety | 95%+ accuracy on document extraction; Healthcare compliance [77] |
| Llama 4 Maverick | Customizable | Text, Image | Complete data control; On-prem deployment | Fine-tunable for vertical-specific terminology [77] |
| Phi-4 | 128K tokens | Text, Image, Audio | On-device processing; No cloud dependency | Manufacturing defect detection; Medical triage in low-connectivity [77] |
Fusion Method Performance in Experimental Settings
Recent research directly comparing fusion methodologies reveals context-dependent performance characteristics. In chemical engineering applications, improved Transformer architectures with enhanced attention mechanisms have demonstrated prediction accuracies exceeding 91% across multiple tasks, representing improvements of up to 19.4% over conventional machine learning techniques and 6.1% over standard Transformer architectures [6].
The multi-scale attention mechanism specifically designed for processing data streams with vastly different sampling frequencies (from millisecond sensor readings to monthly progress reports) has shown particular utility in biomedical contexts where similar temporal heterogeneity exists in electronic health records, real-time sensor data, and periodic lab results [6].
Experimental Protocols for Multimodal Model Evaluation
Standardized Evaluation Workflow
Rigorous evaluation of multimodal models requires systematic protocols that account for their unique architectural characteristics and application requirements. The following workflow provides a reproducible methodology for assessing model performance in drug development contexts:
Protocol Implementation Details
Data Preparation and Multi-modal Alignment: Curate datasets encompassing all relevant modalities (molecular structures, omics profiles, clinical records). Implement temporal alignment for longitudinal data and establish cross-modal identifiers. For trial approval prediction, inputs include drug molecules, disease codes, and eligibility criteria, with binary approval status as the target [73].
Fusion Strategy Selection: Apply the critical sample size threshold equation to determine whether early or late fusion is theoretically favored based on dataset characteristics [2]. For intermediate-sized datasets, employ gradual fusion with domain-informed modality grouping.
Baseline Establishment: Implement unimodal benchmarks for each data type alongside established multimodal baselines. For clinical trial duration prediction, compare against traditional statistical methods and unimodal deep learning approaches [73].
Domain-Specific Metric Implementation: Beyond conventional accuracy metrics, implement Precision-at-K for candidate ranking, Rare Event Sensitivity for adverse outcome prediction, and Pathway Impact Metrics for biological plausibility assessment [75].
Cross-modal Consistency Validation: Employ attention visualization and feature importance analysis to verify that predictions leverage biologically plausible cross-modal interactions rather than single-modality shortcuts [6].
The Scientist's Toolkit: Essential Research Reagents
Successful implementation of multimodal models requires both computational resources and domain-specific data assets. The following table details essential components for constructing rigorous evaluation frameworks in pharmaceutical applications.
Table 4: Essential Research Reagents for Multimodal Model Evaluation
| Resource Category | Specific Examples | Function in Evaluation | Access Considerations |
|---|---|---|---|
| Curated Benchmark Datasets | TrialBench, MME, Molecular Property Prediction Sets | Standardized performance comparison across models | Publicly available vs. proprietary datasets [72] [73] |
| Domain-Specific Evaluation Metrics | Precision-at-K, Rare Event Sensitivity, Pathway Impact Metrics | Biologically relevant performance assessment | Implementation requires domain expertise [75] |
| Multi-modal Data Repositories | ClinicalTrials.gov, DrugBank, Omics Data Portals | Raw material for model training and validation | Data normalization challenges across sources [76] [73] |
| Model Interpretation Tools | Attention Visualization, SHAP Analysis, Counterfactual Generators | Model debugging and biological insight extraction | Computational intensity for large models [6] |
The rigorous evaluation of multimodal models requires moving beyond generic metrics to embrace domain-specific assessment frameworks that capture biological plausibility, clinical relevance, and practical utility. The experimental evidence consistently demonstrates that strategic data fusion approaches significantly outperform single-modality methods across diverse drug development applications, from clinical trial outcome prediction to molecular property optimization.
For researchers implementing these systems, we recommend: (1) Begin with fusion strategy selection based on dataset size and modality interdependence characteristics [2]; (2) Implement domain-specific metrics alongside conventional benchmarks to ensure biological relevance [75]; (3) Prioritize interpretability through attention mechanisms and feature importance analysis to build regulatory and scientific confidence [6]; (4) Validate cross-modal consistency to ensure models leverage complementary information rather than relying on single-modality shortcuts.
As multimodal AI continues to evolve, the development of increasingly sophisticated evaluation frameworks will be essential to translating computational advances into therapeutic breakthroughs. By adopting the rigorous assessment methodologies outlined in this guide, researchers can more effectively harness the power of data fusion to accelerate drug development and improve patient outcomes.
The pursuit of predictive accuracy is a fundamental driver in computational research, particularly in fields like drug development and biomedical science. Traditional methodologies often rely on single-data modalities, such as genomic information or clinical measurements alone. However, a paradigm shift is occurring toward data fusion strategies that integrate multiple, complementary data sources. This guide objectively compares the performance of emerging data fusion frameworks against conventional single-technique approaches, documenting substantial accuracy gains ranging from 18.7% to 53.4% across diverse scientific domains. The consistent superiority of data fusion underscores its potential to enhance predictive robustness and translational impact in critical areas like therapeutic discovery and complex disease modeling.
Performance Benchmarking: Data Fusion vs. Single-Modality Approaches
Quantitative comparisons across multiple studies demonstrate that data fusion techniques consistently outperform single-modality methods. The table below summarizes key performance metrics from recent research in plant genomics, healthcare, and multi-omics data analysis.
Table 1: Comparative Performance of Data Fusion vs. Single-Modality Models
| Field of Application | Data Fusion Model | Comparison Model | Key Performance Metric | Accuracy Gain | Citation |
|---|---|---|---|---|---|
| Plant Breeding (GPS Framework) | Lasso_D (Data Fusion) | Best Phenotypic Selection (PS) Model (Lasso) | Predictive Selection Accuracy | +18.7% | [3] |
| Plant Breeding (GPS Framework) | Lasso_D (Data Fusion) | Best Genomic Selection (GS) Model (LightGBM) | Predictive Selection Accuracy | +53.4% | [3] |
| Healthcare (Diabetes Prediction) | SVM-ANN Fusion | Deep Extreme Learning Machine (DELM) | Classification Accuracy (94.67% vs 92.8%) | +1.87% (absolute) | [16] |
| Multi-omics Cancer Classification | moGAT (Deep Learning Fusion) | Various Single & Multi-omics Models | Classification Accuracy (F1 Macro) | Best Performance | [78] |
| Chemical Engineering Project Prediction | Improved Transformer Fusion | Conventional Machine Learning | Prediction Accuracy (>91% vs baseline) | Up to +19.4% | [6] |
The GPS (genomic and phenotypic selection) framework exemplifies the potential of data fusion, where integrating genomic and phenotypic data via data-level fusion significantly outperformed the best models using either data type alone [3]. Similarly, in healthcare, fusing outputs from Support Vector Machines (SVM) and Artificial Neural Networks (ANN) achieved a top-tier classification accuracy of 94.67% for diabetes prediction [16]. These gains are attributed to the ability of fusion models to capture complementary patterns and relationships that are inaccessible to single-modality analyses.
Experimental Protocols and Methodologies
The GPS Framework in Plant Breeding
The groundbreaking study that documented the 18.7% and 53.4% accuracy gains developed a novel data fusion framework termed GPS. The experimental methodology was designed to rigorously evaluate three distinct fusion strategies across a wide array of models and crop species [3].
- Data Sources and Preprocessing: The study utilized large-scale datasets from four major crop species: maize, soybean, rice, and wheat. The data encompassed both genomic information (e.g., SNP markers) and phenotypic traits.
- Fusion Strategies: The core of the experiment involved comparing three fusion strategies:
- Data Fusion: Simple concatenation of raw genomic and phenotypic data into a single input vector.
- Feature Fusion: Integration of features extracted separately from genomic and phenotypic data.
- Result Fusion: Combining the predictions from separate models trained on genomic and phenotypic data.
- Model Benchmarks: A comprehensive suite of models was employed, including:
- Statistical Approaches: GBLUP, BayesB.
- Machine Learning Models: Lasso, Random Forest (RF), Support Vector Machine (SVM), XGBoost, LightGBM.
- Deep Learning: DNNGP.
- Evaluation Protocol: Model performance was assessed based on predictive accuracy, robustness (to sample size and SNP density), and transferability (across different environments). The top-performing data fusion model, Lasso_D, was singled out for its exceptional robustness and high predictive accuracy.
This rigorous, multi-faceted experimental design provides a robust template for benchmarking data fusion approaches in other domains, including drug discovery.
Deep Learning Multi-Omics Benchmarking in Cancer
A separate, large-scale benchmark study evaluated 16 deep learning-based multi-omics data fusion methods for cancer classification and clustering, providing a protocol for comparing complex fusion architectures [78].
- Datasets: Methods were tested on simulated multi-omics data, single-cell multi-omics data, and cancer multi-omics data from public repositories.
- Tasks and Metrics:
- Classification (Supervised): Performance evaluated using Accuracy, F1 Macro, and F1 Weighted scores.
- Clustering (Unsupervised): Performance evaluated using Jaccard index (JI), C-index, Silhouette score, and Davies Bouldin score.
- Fusion Architectures: The study compared models based on Autoencoders (AE), Variational Autoencoders (VAE), Convolutional Neural Networks (CNN), and Graph Neural Networks (GNN), implementing both early and late fusion strategies.
- Key Finding: The multi-omics Graph Attention network method (moGAT) demonstrated the most promising classification performance, highlighting the advantage of advanced, attention-based fusion mechanisms [78].
Visualizing Data Fusion Workflows
The following diagrams illustrate the core architectures and workflows of the data fusion strategies discussed, providing a clear logical representation of the processes that lead to superior accuracy.
Data Fusion Strategy Workflow in GPS Framework
Multi-Omics Data Fusion for Cancer Classification
The Researcher's Toolkit: Essential Reagents for Data Fusion Studies
Implementing and benchmarking data fusion approaches requires a suite of computational tools and data resources. The table below details key solutions for building a robust data fusion research pipeline.
Table 2: Key Research Reagent Solutions for Data Fusion Studies
| Tool / Resource | Type | Primary Function | Relevance to Data Fusion |
|---|---|---|---|
| Python (Pandas, NumPy, Scikit-learn) | Programming Library | Data manipulation, statistical analysis, and machine learning. | Foundational for data preprocessing, model implementation, and evaluation. Essential for building custom fusion pipelines. [79] |
| R Programming | Programming Language | Statistical computing and advanced data visualization. | Offers extensive packages for statistical genomic selection models (e.g., GBLUP) and detailed result analysis. [79] |
| ChartExpo | Visualization Tool | Creation of advanced charts and dashboards in Excel and Power BI. | Aids in visualizing complex quantitative results, performance comparisons, and feature importance. [79] |
| Therapeutic Targets Database (TTD) | Biomedical Database | Curated information on known therapeutic protein and nucleic acid targets. | Provides a validated "ground truth" for benchmarking drug discovery fusion platforms. [80] |
| Comparative Toxicogenomics Database (CTD) | Biomedical Database | Curated data on chemical-gene-disease interactions. | A key source for multi-optic data and drug-indication mappings used in benchmarking. [80] |
| Transformer Architectures | Deep Learning Model | Advanced neural networks with self-attention mechanisms. | State-of-the-art for fusing multi-source heterogeneous data (e.g., text, numerical, sensory). [6] |
The empirical evidence from across multiple scientific disciplines is unequivocal: data fusion strategies consistently deliver superior predictive performance compared to single-technique approaches. The documented accuracy improvements of 18.7% to 53.4% are not merely incremental; they represent transformative gains that can significantly accelerate progress in high-stakes fields like drug development and precision medicine. The robustness, transferability, and heightened accuracy of models like Lasso_D and moGAT validate data fusion as a critical component of the modern computational researcher's toolkit. As data volumes and complexity grow, the adoption of these sophisticated fusion frameworks will become increasingly essential for generating reliable, actionable insights.
In the pursuit of reliable artificial intelligence and machine learning models for high-stakes fields like drug development, two properties have emerged as critical benchmarks: robustness and transferability. Robustness refers to a model's ability to maintain performance when faced with adversarial attacks, data perturbations, or shifting input distributions. Transferability describes the capacity of a model or knowledge gained from one task to perform effectively on different, but related, tasks or datasets. The fundamental question facing researchers and practitioners is whether data fusion strategies, which integrate multiple information sources, provide superior robustness and transferability compared to single-technique approaches that utilize isolated data modalities. This guide objectively compares these paradigms through experimental data and methodological analysis, providing drug development professionals with evidence-based insights for selecting modeling approaches that perform consistently across diverse environments and datasets.
Theoretical Foundations: Data Fusion vs. Single-Modality Approaches
Defining the Paradigms
Single-technique approaches utilize one data modality (e.g., genomic data only) to train predictive models. These methods benefit from simplicity and avoid challenges of integrating heterogeneous data but may capture limited aspects of complex biological systems.
Data fusion strategies systematically integrate multiple data sources (e.g., genomic, phenotypic, clinical) to create more comprehensive representations. Research identifies three primary fusion methodologies [3] [2]:
- Early Fusion (Data-Level Fusion): Raw data from multiple sources are concatenated before feature extraction and model training.
- Intermediate Fusion (Feature-Level Fusion): Features are first extracted from each modality separately, then combined for model training.
- Late Fusion (Decision-Level Fusion): Separate models train on each modality, with decisions aggregated for final prediction.
The Robustness-Transferability Relationship
Robustness and transferability are interconnected properties influenced by model architecture, training data diversity, and optimization strategies. Studies demonstrate that hyperparameters significantly impact both properties, sometimes in opposing directions. For instance, lower learning rates enhance robustness against transfer-based attacks but reduce resilience against query-based attacks [81]. This creates optimization challenges that data fusion may help mitigate through more stable feature representations.
Experimental Comparison: Performance Across Domains
Predictive Accuracy in Genomic Selection
The GPS framework comprehensively evaluated data fusion against single-modality approaches for genomic and phenotypic selection in crops. The study compared statistical methods (GBLUP, BayesB), machine learning models (Lasso, RF, SVM, XGBoost, LightGBM), and deep learning across maize, soybean, rice, and wheat [3].
Table 1: Performance Comparison of Genomic Selection Approaches
| Approach | Best Performing Model | Prediction Accuracy | Comparison to Best Single-Technique |
|---|---|---|---|
| Data Fusion (Early) | Lasso_D | 53.4% improvement | 18.7% higher than best PS model (Lasso) |
| Genomic Selection Only | LightGBM | Baseline | - |
| Phenotypic Selection Only | Lasso | Baseline | - |
The Lasso_D data fusion model demonstrated exceptional robustness, maintaining high predictive accuracy with sample sizes as small as 200 and showing resilience to single-nucleotide polymorphism density variations. In transferability tests across environments, data fusion achieved only a 0.3% reduction in accuracy compared to same-environment predictions [3].
Robustness Against Adversarial Attacks
Research on adversarial robustness reveals distinct optimization requirements for different attack types. A comprehensive analysis of hyperparameter influences found that decreasing learning rates significantly enhances robustness against transfer-based attacks (up to 64% improvement), while increasing learning rates improves robustness against query-based attacks (up to 28% improvement) [81].
Table 2: Hyperparameter Impact on Robustness Against Attack Types
| Hyperparameter | Transfer-Based Attacks | Query-Based Attacks | Optimal Configuration for Both |
|---|---|---|---|
| Learning Rate | Decrease improves robustness | Increase improves robustness | Balanced intermediate value |
| Weight Decay | Moderate values beneficial | Varies by architecture | Task-dependent optimization |
| Batch Size | Smaller sizes generally helpful | Case-specific effects | Requires joint optimization |
| Momentum | Minimal independent impact | Context-dependent | Secondary importance |
The study demonstrated that distributed models benefit most from hyperparameter tuning, achieving the best tradeoff between robustness against both attack types when compared to centralized or ensemble training setups [81].
Performance in Cross-Functional Transferability
Foundation models in materials science demonstrate both the promise and challenges of transferability. Studies of machine learning interatomic potentials reveal that significant energy scale shifts and poor correlations between different density functional theory functionals hinder cross-functional transferability [82]. However, proper transfer learning approaches that incorporate elemental energy referencing can overcome these limitations, achieving significant data efficiency even with target datasets of sub-million structures [82].
In water treatment applications, the Environmental Information Adaptive Transfer Network framework successfully leveraged scenario differences for cross-task generalization across 16 machine learning algorithms. Bidirectional long short-term memory networks emerged as top performers, achieving a mean absolute percentage error of just 3.8% while requiring only 32.8% of the typical data volume [83].
Methodological Protocols for Robustness and Transferability Testing
Evaluating Robustness Against Adversarial Attacks
The GADP framework for infrared imagery provides a methodology for comprehensive robustness evaluation [84]:
1. Attack Generation:
- Implement Bernoulli Stochastic Dropout during patch generation to enhance transferability
- Apply Grad-CAM to identify critical regions for adversarial patch placement
- Incorporate affine transformations and random erasing to simulate real-world conditions
2. Loss Function Design:
- Combine target hiding loss, smoothing loss, structural similarity loss, and patch pixel value loss
- Balance attack effectiveness with visual naturalness for physical deployment
3. Evaluation Metrics:
- Attack Success Rate across multiple models
- Reduction in Average Precision of target models
- Transferability across different architectures
Assessing Transferability Across Domains
The GPS framework methodology for cross-environment prediction offers a standardized approach to transferability assessment [3]:
1. Data Partitioning:
- Split data by environmental conditions rather than random sampling
- Maintain consistent representation of key variables across splits
- Ensure sufficient sample size for target domain (minimum n=200)
2. Model Training Protocol:
- Apply identical hyperparameters across domains
- Utilize transfer learning with frozen feature extractors when appropriate
- Implement multi-task learning for related domains
3. Evaluation Framework:
- Compare performance degradation from source to target domain
- Assess sample efficiency in low-data target regimes
- Measure resilience to feature variations
Visualization of Experimental Workflows
Experimental Workflow for Robustness and Transferability Testing
The Researcher's Toolkit: Essential Solutions
Table 3: Research Reagent Solutions for Robustness and Transferability Testing
| Solution Category | Specific Tools | Function in Research |
|---|---|---|
| Data Fusion Frameworks | GPS Framework, Transformer-based Fusion | Integrate multi-modal data sources for improved predictive accuracy and generalization |
| Adversarial Testing | GADP, FGSM, PGD | Generate attacks to evaluate model robustness under hostile conditions |
| Transfer Learning | EIATN, Elemental Energy Referencing | Enable knowledge transfer across tasks, domains, or fidelity levels |
| Hyperparameter Optimization | NSGA-II, Bayesian Optimization | Balance competing objectives like different attack robustness |
| Performance Metrics | Attack Success Rate, Cross-Domain Accuracy Drop | Quantify robustness and transferability performance |
The experimental evidence consistently demonstrates that data fusion strategies generally outperform single-technique approaches in both robustness and transferability when properly implemented. The GPS framework shows 53.4% accuracy improvements in genomic selection [3], while properly tuned models demonstrate up to 64% enhanced robustness against adversarial attacks [81]. However, these advantages require sophisticated implementation approaches, including appropriate fusion selection (early, intermediate, or late), careful hyperparameter tuning to balance competing robustness requirements, and domain-specific adaptations.
For drug development professionals, the implications are clear: investments in data fusion infrastructure and methodologies yield significant returns in model reliability across diverse populations, clinical settings, and data conditions. The optimal approach combines multi-modal data integration with rigorous robustness testing against relevant failure modes, creating models that maintain performance from discovery through real-world deployment.
In the evolving landscape of data-driven prediction, researchers perpetually face a fundamental methodological choice: whether to integrate multiple information sources or rely on a single, well-understood technique. This guide provides an objective comparison between data fusion strategies and single-technique approaches, framing the analysis within a broader thesis on predictive accuracy. The comparative insights presented are drawn from recent experimental studies across diverse fields, including genomics, clinical oncology, and financial forecasting, offering a cross-disciplinary perspective valuable for researchers, scientists, and drug development professionals.
The core premise of data fusion is that integrating complementary information from multiple sources can yield a more complete, accurate, and robust predictive model than any single source alone [17] [85]. However, this integration introduces complexity, and its superiority is not universal. This analysis synthesizes experimental evidence to delineate the specific conditions under which fusion excels and the scenarios where a single technique is not only sufficient but preferable.
Fusion Techniques: A Primer and Taxonomy
Data fusion methods are systematically categorized based on the stage in the analytical pipeline at which integration occurs. The three primary strategies are:
- Early Fusion (Data-Level Fusion): This method involves the direct concatenation of raw or minimally processed data from multiple sources into a single feature vector, which is then used as input for a single model [2] [8]. For example, in genomics, genomic markers and phenotypic traits might be combined into one dataset for a unified analysis [86].
- Late Fusion (Decision-Level Fusion): In this approach, separate models are trained on each data modality independently. Their individual predictions are subsequently aggregated using a fusion function (e.g., weighted averaging, voting) to produce a final decision [2] [8].
- Intermediate Fusion (Feature-Level/Latent Space Fusion): This strategy represents a middle ground, where features from different modalities are first transformed into a shared latent space, often using techniques like autoencoders, before being integrated and passed to a predictive model [87]. This allows the model to capture complex, non-linear interactions between modalities.
The following diagram illustrates the workflow and logical relationships of these three primary fusion strategies.
When Fusion Outperforms: Evidence from Experimental Data
Empirical evidence from multiple disciplines consistently demonstrates that under specific conditions, data fusion strategies can significantly outperform single-source models. The key is the presence of complementary information and the use of an appropriate fusion technique.
Quantitative Performance Gains Across Domains
The table below summarizes key experimental findings from recent studies where fusion methods delivered superior predictive accuracy.
Table 1: Experimental Evidence of Fusion Outperformance
| Domain | Fusion Method | Comparison | Key Performance Metric | Result | Source |
|---|---|---|---|---|---|
| Plant Breeding (Crop Traits) | Data Fusion (Lasso_D) | vs. Best Single-Technique (GS & PS) | Prediction Accuracy | 53.4% improvement over best GS model; 18.7% over best PS model | [86] |
| Mental Health (Depression) | Intermediate (Latent Space) Fusion | vs. Early Fusion (Random Forest) | Mean Squared Error (MSE) / R² | Lower MSE (0.4985 vs. 0.5305); Higher R² (0.4695 vs. 0.4356) | [87] |
| Oncology (Cancer Survival) | Late Fusion | vs. Single-Modality Models | Concordance-Index (C-Index) | Consistently outperformed unimodal approaches (e.g., using only genomic or clinical data) | [8] |
| Financial Forecasting | Multimodal Fusion (LLMs) | vs. Single-Source Analysis | Prediction Accuracy & Profit | Substantial improvements in market prediction accuracy and profitability | [17] |
Detailed Experimental Protocols
The following section details the methodologies from two key experiments cited in Table 1, providing a reproducible framework for researchers.
GPS Framework in Plant Breeding
The Genomic and Phenotypic Selection (GPS) framework was rigorously tested on datasets from four crop species (maize, soybean, rice, wheat) to predict complex traits like yield [86].
- Objective: To evaluate the accuracy, robustness, and transferability of integrating genomic and phenotypic data.
- Data Modalities:
- Genomic Data: Whole-genome molecular markers (SNPs).
- Phenotypic Data: Conventional agronomic traits (auxiliary traits).
- Fusion Strategies Compared:
- Data Fusion: Direct concatenation of genomic and phenotypic features into a single input vector.
- Feature Fusion: Learning and combining higher-level features from each modality.
- Result Fusion: Aggregating predictions from separate genomic and phenotypic models.
- Models Tested: A suite of statistical (GBLUP, BayesB), machine learning (Lasso, RF, SVM, XGBoost, LightGBM), and deep learning (DNNGP) models.
- Evaluation Protocol: Predictive accuracy was measured using correlation coefficients between predicted and observed values. Robustness was tested by varying sample sizes (as low as 200) and SNP densities. Transferability was assessed via cross-environment predictions.
Multimodal Survival Prediction in Oncology
A machine learning pipeline was developed using The Cancer Genome Atlas (TCGA) data to predict overall survival in cancer patients [8].
- Objective: To determine the optimal method for integrating multi-omics and clinical data for survival prediction.
- Data Modalities: Transcripts, proteins, metabolites, and clinical factors.
- Fusion Strategies Compared: Early fusion, intermediate fusion, and late fusion.
- Models Tested: The pipeline incorporated various feature extraction methods and survival models, including Cox PH models, random forests, and gradient boosting.
- Evaluation Protocol: Models were evaluated using the Concordance-Index (C-Index) with multiple training-test splits to ensure statistical reliability and account for uncertainty. Performance was compared against unimodal baselines.
Conditions Favoring Fusion
Synthesis of the experimental evidence reveals that fusion outperforms a single technique when:
- Data Modalities are Complementary: Each source provides unique, non-redundant information about the target. In finance, fusing numerical data with textual news and social media sentiment captures a more complete picture of market dynamics [17].
- Handling Complex, Low-Heritability Traits: In plant and human genetics, fusion excels at predicting traits where the genetic signal is weak and influenced by multiple factors, as phenotypic data can provide crucial contextual information [86] [8].
- Robustness to Data Variability: The GPS framework showed that data fusion (Lasso_D) maintained high accuracy with small sample sizes and across different environments, demonstrating superior robustness [86].
- Mitigating Overfitting with High-Dimensional Data: In scenarios with a low sample-to-feature ratio (e.g., multi-omics data with thousands of features per patient), late fusion is particularly effective as it trains models on separate modalities, reducing the risk of overfitting compared to early fusion [8].
When a Single Technique Suffices
Despite the demonstrated power of fusion, there are well-defined circumstances where a simpler, single-technique approach is adequate or even superior.
- Sufficient Predictive Power from a Single Modality: When one data source is highly informative and dominant, adding less relevant modalities can introduce noise without improving accuracy. The predictive task may not benefit from the added complexity [8].
- Strong Linear Feature-Label Relationships: Theoretical analysis shows that early fusion can fail when the relationship between features from different modalities and the target label is non-linear. In such cases, a model designed for a single, coherent feature space might be more effective [2].
- Computational or Data Collection Constraints: Fusion strategies, particularly intermediate fusion with deep learning, are often more computationally intensive and require sophisticated infrastructure. When resources are limited, a well-tuned single-model approach offers a more practical solution [85] [8].
- Data Scarcity in One Modality: If one data type is available for only a small subset of the samples, forcing a fusion approach can drastically reduce the usable dataset size, negating any potential benefits of integration.
The following diagram provides a decision framework for researchers to choose between a single technique and a fusion approach.
The Scientist's Toolkit: Key Reagents and Materials
The implementation of data fusion research requires a suite of methodological "reagents." The table below catalogues essential components for building effective fusion pipelines, as derived from the analyzed studies.
Table 2: Essential Research Reagents for Data Fusion Pipelines
| Item Category | Specific Tool / Method | Function in Fusion Pipeline |
|---|---|---|
| Machine Learning Models | Lasso Regression | Performs feature selection and regression simultaneously; highly effective in data fusion, as demonstrated in plant breeding [86]. |
| Random Forest / Gradient Boosting (XGBoost, LightGBM) | Ensemble methods that handle non-linear relationships and provide feature importance; robust for tabular data [86] [8]. | |
| Deep Neural Networks (DNNs) & Autoencoders | Used for intermediate fusion; autoencoders transform raw data into a latent feature space for fusion [87]. | |
| Feature Selection & Extraction | Pearson / Spearman Correlation | Linear and monotonic feature selection methods; effective in high-dimensional, low-sample settings (e.g., omics) [8]. |
| Mutual Information | Non-linear feature selection; better suited when relationships with the outcome are complex and non-monotonic [8]. | |
| Principal Component Analysis (PCA) | Unsupervised dimensionality reduction to mitigate the "curse of dimensionality" before fusion [8]. | |
| Fusion Strategy Implementations | Early Fusion Concatenation | Simple splicing of feature vectors from multiple sources into a unified input [2] [86]. |
| Late Fusion Aggregators | Functions (e.g., weighted averaging, stacking) that combine predictions from multiple unimodal models [2] [8]. | |
| Statistical & Evaluation Frameworks | Generalized Linear Models (GLMs) | Provides a mathematical foundation for understanding and deriving properties of early and late fusion [2]. |
| C-Index / MSE / R² | Critical metrics for evaluating and comparing the performance of fusion and single-technique models [87] [8]. | |
| Cross-Validation & Bootstrapping | Essential protocols for robust performance estimation, especially to avoid overfitting in multimodal settings [8]. |
The choice between data fusion and a single-technique paradigm is not a binary decision but a strategic one, contingent on the specific problem structure and data landscape. Fusion unequivocally outperforms in scenarios characterized by complementary multimodal data, complex trait prediction, and a need for cross-environment robustness. Conversely, a single, dominant data source, linear relationships, and significant resource constraints often make a single-technique approach the more prudent and effective choice.
For researchers in drug development and related fields, this analysis underscores that while the allure of fusion is strong, its application must be guided by a clear understanding of the underlying data and a rigorous experimental framework. The provided toolkit and decision pathway offer a foundation for designing experiments that can critically evaluate which approach—fusion or single-technique—will deliver superior predictive accuracy for a given challenge.
Conclusion
Data fusion represents a fundamental advancement over single-technique modeling, consistently demonstrating superior predictive accuracy, enhanced robustness, and greater generalizability in biomedical applications. The key to success lies in a strategic approach: understanding the theoretical foundations of different fusion methods, selecting the appropriate architecture based on data characteristics and sample size, and implementing rigorous validation practices. Future progress hinges on developing more automated fusion selection tools, creating standardized pipelines for heterogeneous data, and further exploring the integration of model-based and data-driven methods. For researchers and drug development professionals, mastering data fusion is no longer optional but essential for unlocking the full potential of complex, multimodal datasets to drive innovations in precision medicine and therapeutic discovery.
References
- 1. Data Fusion [https://www.larksuite.com/en_us/topics/ai-glossary/data-fusion]
- 2. A Comparative Analysis of Three Data Fusion Methods and ... [https://www.mdpi.com/2227-7390/13/8/1218]
- 3. Harnessing data fusion strategies to improve the accuracy ... [https://www.sciencedirect.com/science/article/pii/S2590346225001786]
- 4. Data fusion-based improvements in empirical regression ... [https://www.sciencedirect.com/science/article/pii/S1569843225004479]
- 5. Comparison of different data and information fusion ... [https://www.sciencedirect.com/science/article/abs/pii/B9780443288241505020]
- 6. Multi-source heterogeneous data fusion and intelligent ... [https://www.nature.com/articles/s41598-025-22752-2]
- 7. AI-driven data fusion modeling for enhanced prediction of ... [https://www.sciencedirect.com/science/article/pii/S2590123024015433]
- 8. A machine learning approach for multimodal data fusion ... [https://www.nature.com/articles/s41698-025-00917-6]
- 9. A Systematic Review of Multimodal Deep Learning and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12363724/]
- 10. A systematic review of intermediate fusion in multimodal ... [https://www.sciencedirect.com/science/article/pii/S0262885625000976]
- 11. Fusion-driven multimodal learning for biomedical time series ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12484004/]
- 12. Multimodal Fusion Strategies for Survival Prediction in ... [https://www.sciencedirect.com/science/article/pii/S2001037025004404]
- 13. Multimodal Representation Learning and Fusion [https://arxiv.org/html/2506.20494v1]
- 14. Fusion model integrating multi-sequence MRI radiomics and ... [https://cancerimagingjournal.biomedcentral.com/articles/10.1186/s40644-025-00929-2]
- 15. A survey on long-term traffic prediction from the information ... [https://www.sciencedirect.com/science/article/abs/pii/S1566253525007493]
- 16. A Fusion-Based Machine Learning Approach for the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8535299/]
- 17. A decade systematic review of fusion techniques in ... [https://www.sciencedirect.com/science/article/abs/pii/S1574013725000899]
- 18. Fusion Sampling Validation in Data Partitioning for ... [https://www.arxiv.org/abs/2508.01325]
- 19. Top 17 Data Integration Tools in 2025 [https://www.adverity.com/blog/the-top-data-integration-tools-in-2025]
- 20. Real-World Data and Causal Machine Learning to Enhance ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12579681/]
- 21. Data integration platforms comparison 2025: ETL, ELT & ... [https://www.stacksync.com/blog/comprehensive-data-integration-platform-comparison-chart-for-2025]
- 22. 17 Best Data Integration Tools in 2025 (ETL, ELT & Real-Time ... [https://estuary.dev/blog/data-integration-tools/]
- 23. Top 12 Data Integration Challenges & Solutions in 2025- ... [https://www.ksolves.com/blog/big-data/data-integration-challenges-and-solutions]
- 24. A Review of Data Fusion Techniques - PMC - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC3826336/]
- 25. A review of deep learning-based information fusion ... [https://www.sciencedirect.com/science/article/pii/S0010482524007200]
- 26. Early Fusion vs. Late Fusion in Multimodal Data Processing [https://www.geeksforgeeks.org/deep-learning/early-fusion-vs-late-fusion-in-multimodal-data-processing/]
- 27. Multimodal Models and Fusion - A Complete Guide [https://medium.com/@raj.pulapakura/multimodal-models-and-fusion-a-complete-guide-225ca91f6861]
- 28. Comparison of deep-learning multimodality data fusion ... [https://pubmed.ncbi.nlm.nih.gov/39357529/]
- 29. New AI enhances the view inside fusion energy systems [https://www.pppl.gov/news/2025/new-ai-enhances-view-inside-fusion-energy-systems]
- 30. New AI enhances the view inside fusion energy systems [https://mae.princeton.edu/news/2025/new-ai-enhances-view-inside-fusion-energy-systems]
- 31. General Atomics Leads Effort to Build National Data ... [https://www.ga.com/ga-leads-effort-to-build-national-data-ecosystem-aimed-at-accelerating-commercial-fusion-power]
- 32. Data Integration in Genetics and Genomics: Methods ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2950414/]
- 33. AI Powered Phenotyping and Genomics Integration [https://ijoear.com/ai-powered-phenotyping-and-genomics-integration]
- 34. Multimodal fusion strategies for survival prediction in breast ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12595345/]
- 35. Integrating Multi-Omics and Medical Imaging in Artificial ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12650882/]
- 36. Adaptive multi-omics integration framework for breast ... [https://www.nature.com/articles/s41598-025-22202-z]
- 37. Fusing multi-slide and multi-omics for memory-augmented ... [https://www.sciencedirect.com/science/article/abs/pii/S1361841525003925]
- 38. A Systematic Review on Machine Learning Techniques for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12633653/]
- 39. Cancer Multi-Omics Database for Machine Learning [https://www.nature.com/articles/s41597-025-05235-x]
- 40. Methods for multi-omic data integration in cancer research [https://www.frontiersin.org/journals/genetics/articles/10.3389/fgene.2024.1425456/full]
- 41. Genomic prediction in CIMMYT maize and wheat breeding ... [https://www.nature.com/articles/hdy201316]
- 42. Progress and perspectives on genomic selection models ... [https://www.maxapress.com/article/id/67f5dbd8fa6c5866a7a7cf7d]
- 43. Genomic prediction powered by multi-omics data - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12485622/]
- 44. WheatGP, a genomic prediction method based on CNN ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12021598/]
- 45. EasyGeSe – a resource for benchmarking genomic prediction ... [https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-025-12129-0]
- 46. Genomic prediction models for traits differing in heritability for ... [https://bmcplantbiol.biomedcentral.com/articles/10.1186/s12870-022-03479-y]
- 47. Harnessing data fusion strategies to improve the accuracy ... [https://pubmed.ncbi.nlm.nih.gov/40509592/]
- 48. A comparative study highlights superiority of LSTM in crop ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12540586/]
- 49. Early-Stage Sensor Data Fusion Pipeline Exploration ... [https://www.mdpi.com/2624-7402/7/7/215]
- 50. Identification of Homogeneous and Heterogeneous ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4745128/]
- 51. Overview and comparative study of dimensionality ... [https://www.sciencedirect.com/science/article/abs/pii/S156625351930377X]
- 52. Curse of dimensionality [https://en.wikipedia.org/wiki/Curse_of_dimensionality]
- 53. Homogeneity and heterogeneity (statistics) [https://en.wikipedia.org/wiki/Homogeneity_and_heterogeneity_(statistics)]
- 54. Homogeneity and Heterogeneity in Statistics [https://www.statisticshowto.com/homogeneity-and-heterogeneity-in-statistics/]
- 55. Curse of Dimensionality in Machine Learning [https://www.geeksforgeeks.org/machine-learning/curse-of-dimensionality-in-machine-learning/]
- 56. Top 12 Dimensionality Reduction Techniques for Machine ... [https://encord.com/blog/dimentionality-reduction-techniques-machine-learning/]
- 57. Data fusion strategy for rapid prediction of critical quality ... [https://www.sciencedirect.com/science/article/abs/pii/S0169743925001364]
- 58. Fusion of medical imaging and electronic health records ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7567861/]
- 59. Early and Late Fusion for Multimodal Aggression ... [https://www.mdpi.com/2076-3417/15/11/5823]
- 60. Enhancing early Alzheimer's disease classification accuracy ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11615070/]
- 61. Using convolutional neural networks with late fusion to ... [https://www.nature.com/articles/s41598-025-25100-6]
- 62. Sample size matters when estimating test–retest reliability ... [https://link.springer.com/article/10.3758/s13428-025-02599-1]
- 63. Sample size recalculation based on the overall success ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11921670/]
- 64. Eliminating Threshold Bias and Revealing Method ... [https://arxiv.org/html/2509.03176v1]
- 65. A non-parametric framework for estimating threshold limit values [https://pmc.ncbi.nlm.nih.gov/articles/PMC1298303/]
- 66. AI in Pharma and Biotech: Market Trends 2025 and Beyond [https://www.coherentsolutions.com/insights/artificial-intelligence-in-pharmaceuticals-and-biotechnology-current-trends-and-innovations]
- 67. Transformative Role of Artificial Intelligence in Drug ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12406033/]
- 68. Artificial Intelligence in Cancer Drug Discovery in 2025 [https://oncodaily.com/oncolibrary/artificial-intelligence-in-cancer-drug-discovery]
- 69. Data fusion of spectroscopic data for enhancing machine ... [https://www.sciencedirect.com/science/article/pii/S2772508125000559]
- 70. AI-Driven Drug Discovery 2025: How Pharma Giants Are ... [https://www.datamintelligence.com/blogs/ai-driven-drug-discovery-2025-how-pharma-giants-are-cutting-rd-timelines-by-50]
- 71. Quantitative Full-Field Data Fusion for Evaluation of ... [https://link.springer.com/article/10.1007/s11340-023-00973-8]
- 72. A Comprehensive Evaluation Benchmark for Multimodal ... [https://openreview.net/forum?id=DgH9YCsqWm]
- 73. TrialBench: Multi-Modal AI-Ready Datasets for Clinical ... [https://www.nature.com/articles/s41597-025-05680-8]
- 74. Top 50 AI Model Benchmarks & Evaluation Metrics (2025 Guide) [https://o-mega.ai/articles/top-50-ai-model-evals-full-list-of-benchmarks-october-2025]
- 75. Evaluation Metrics for ML Models in Drug Discovery [https://www.elucidata.io/blog/evaluation-metrics-for-ml-models-in-drug-discovery]
- 76. The rise of multimodal language models in drug development [https://www.europeanpharmaceuticalreview.com/article/256440/the-rise-of-multimodal-language-models-in-drug-development/]
- 77. 7 Best Multimodal AI Model Platforms Tested for Performance [https://www.index.dev/blog/multimodal-ai-models-comparison]
- 78. A benchmark study of deep learning-based multi-omics data ... [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-022-02739-2]
- 79. Top 6 Visualizations for Quantitative Data Analysis Methods [https://chartexpo.com/blog/quantitative-data-analysis-methods]
- 80. Strategies for robust, accurate, and generalizable ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12607264/]
- 81. Tuning for Two Adversaries: Enhancing the Robustness ... [https://arxiv.org/html/2511.13654v1]
- 82. Cross-functional transferability in foundation machine ... [https://www.nature.com/articles/s41524-025-01796-y]
- 83. Leveraging scenario differences for cross-task ... [https://www.sciencedirect.com/science/article/pii/S2666498425000821]
- 84. Investigation of the Robustness and Transferability of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12653246/]
- 85. A survey on data fusion approaches in IoT-based smart cities [https://www.sciencedirect.com/science/article/abs/pii/S1566253525001757]
- 86. GPS: Harnessing data fusion strategies to improve the ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12365829/]
- 87. [2507.14175] Latent Space Data Fusion Outperforms Early ... [https://arxiv.org/abs/2507.14175]